– Our database is accessible HERE, or through our CloneMap tool.
Highlights
– A collection of human ORFs, V5-tagged at their C-terminus, and expression-ready in a lentiviral vector plasmid.
– Expression can be achieved both by simple transfection of the plasmid, or by lentiviral-mediated transduction. In both cases, only an insignificant amount of lentiviral vectors-derived elements is expressed along with the ORF.
– ORFs are under the control of a CMV promoter, and are devoid of native UTRs.
– ORFs can easily be shuttled towards other expression plasmids owing to the Gateway technology.
– The collection contains approximately 13’000 ORFs, representing ca. 11’000 unique protein-coding genes.
In more details
– The collection was created by teams of the CCSB-Dana Farber Institute and the Broad Institute, by shuttling the hORFeome v8.1 library towards a lentiviral destination vector in 2011 (original paper). As for the ORFeome Collaboration (OC), the initial parental cDNAs were mostly derived from the Mammalian Gene Collection (MGC).
– If you don’t want your ORF of interest in a lentiviral vector system, or if you don’t want a V5 tag, the ORF (untagged, without STOP codon) can be shuttled by Gateway techniques towards other Gateway destination vectors, with just an intermediary step. The restriction site-independence and high-fidelity of this method allows for the batch shuttling of large subsets of clones in parallel. Alternatively, ORFs can be subcloned as desired by standard PCR-based cloning.
– The CMV promoter is generally broadly active but may not be suited in certain type of cells or organisms. In case of doubt, please Google or contact us.
– No native UTRs are present in these clone, but a Kozak-like sequence is.
– Transduced cells can be selected by Blasticidin.
– Control vectors carrying a LacZ transgene or no transgene at all are available upon request.
– While its parental library was sequenced in great part (see below), the final CCSB-Broad lentiviral Expression library was not sequenced by their creators. Indeed, the Gateway technology being non-PCR-mediated, cloning errors are expected to have been very rare during the shuttling from the parental library to this final library. Nevertheless, we highly recommend sequencing of the clones you receive from us. When a clone has already been fully sequenced at GECF, it is indicated in our database.
– The parental hORFeome v8.1 collection was sequenced in great part. During this sequencing process, it was noted that a fraction of the clones contains mutations, likely PCR-generated. These clones were assembled into a subcollection named “mutant subcollection”. In addition some clones were not fully sequenced and were grouped accordingly in the subcollection named “unsequenced subcollection”. We indicate in our Excel database which clones belong to these subcollections. To obtain the exact sequence of these clones, follow the procedure here, or simply contact us.
– The presence in this collection of ORFs with oncogenic potential (upon lentivector-mediated overexpression or ectopic expression) deserves an extra-level of care. Consider working with BSL2+ safety conditions (BSL3-like procedures in a BSL2 laboratory) when needed.
References for a publication
When using a lentiviral vector in a publication, refer to it as part of the “CCSB-Broad lentiviral expression library”, and reference this publication: https://www.nature.com/articles/nmeth.1638. The ID that should be indicated is the “IMAGE ID” or “Original ID” from our Excel database/CloneMap.
MTA
– There is no restriction to the use of the clones and their derivatives for non-commercial application. Nevertheless, transferring the clones outside of EPFL requires that the original MTA be included and that it is done free of charge. This requirement does not apply to derivatives of the clones (ORF subcloned in another vector for instance).
– If commercial applications are foreseen, please contact us for more details, in particular regarding the Gateway elements.
– The MTA can be found HERE.
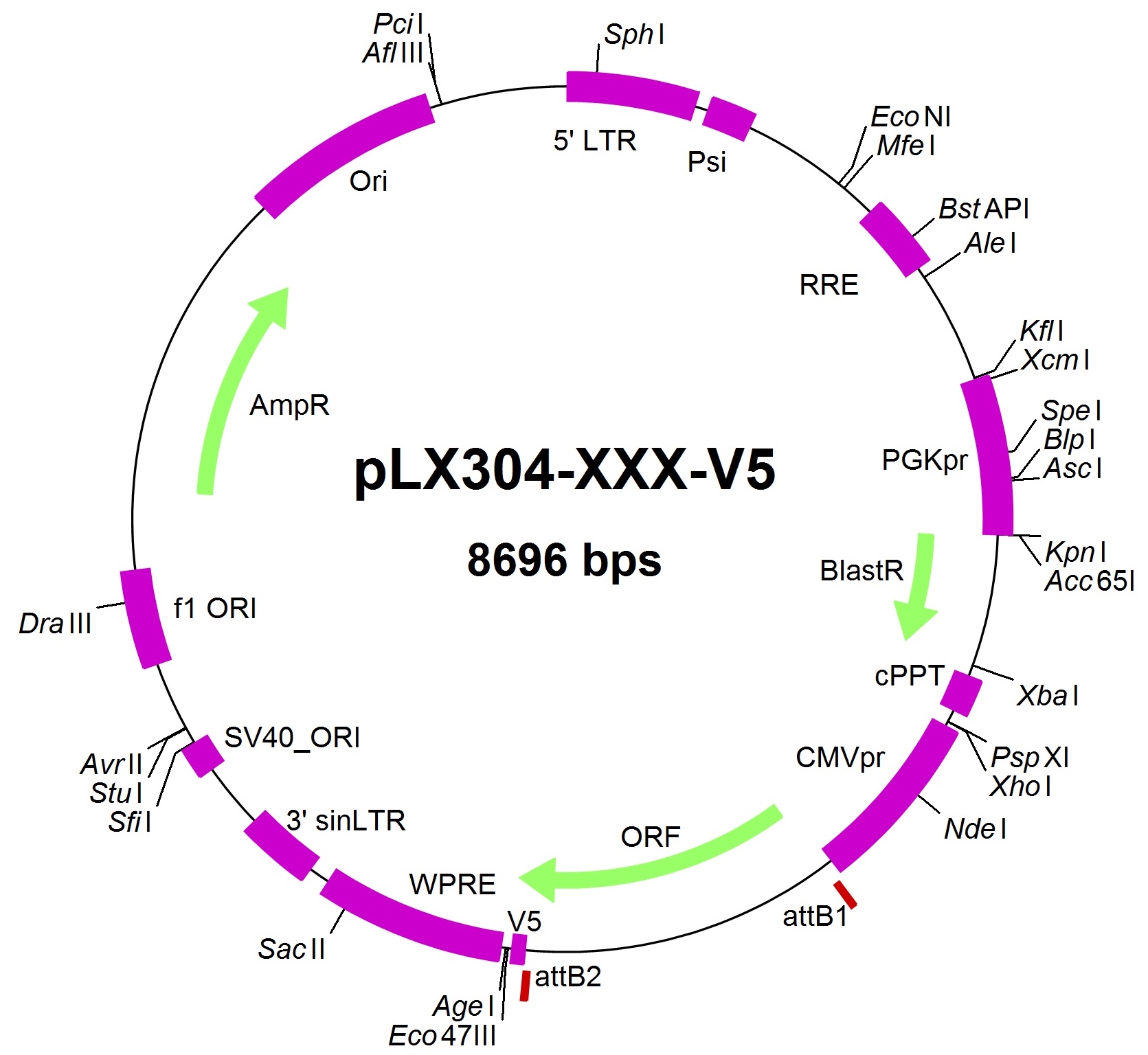
Maps
– The maps are available in our CloneMap tool.
– The sequence of the lentiviral vector containing a virtual ORF is available HERE, and its sketch is here: