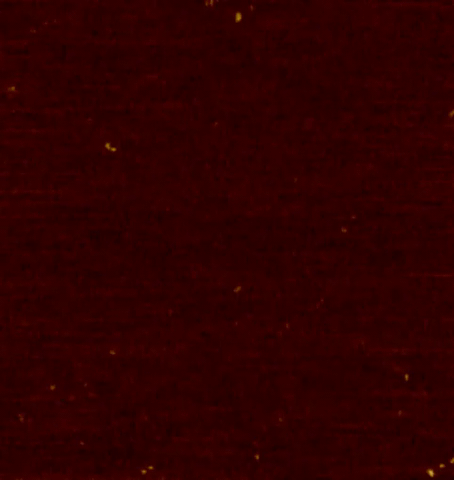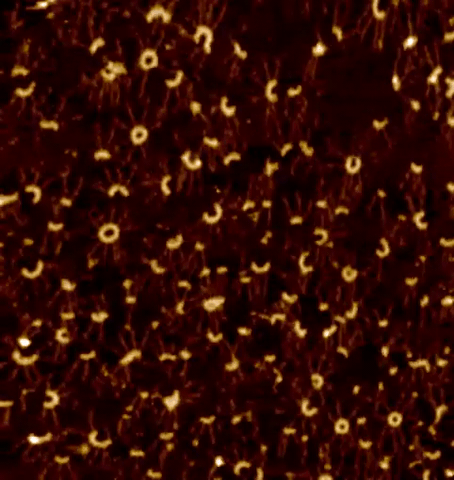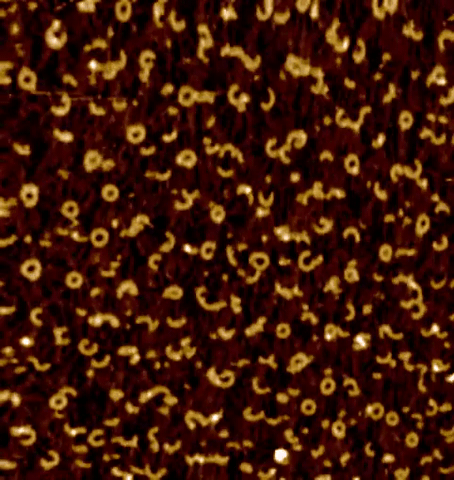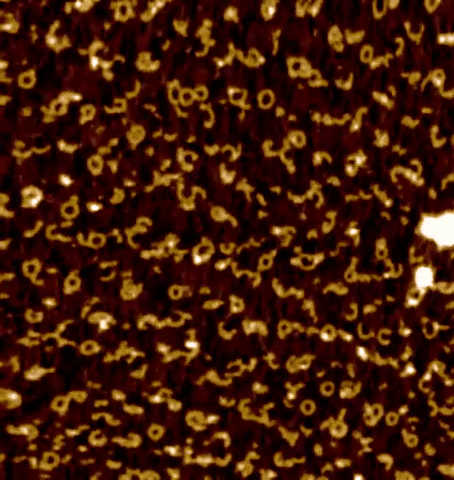
Centrioles are nine-fold radially symmetric microtubule-based organelles that play critical roles in eukaryotic cell physiology. Centriole assembly requires scaffold protein, homodimers of SAS-6, undergo a higher order oligomerization through a weak interaction mediated by their globular head domain. These higher-order oligomers form nine-fold symmetric ring-like structures that initiate centriole formation in vivo and impart the signature nine-fold radial symmetry to the entire organelle. However, the dynamics of the SAS-6 ring assembly have not been resolved to date, which precludes a thorough understanding of the mechanisms at the root of centriole biogenesis. Using HS-PORT allowed us to monitor the molecular dynamics of SAS-6 self-assembly.
The formation of lethal membrane pores is a ubiquitous event in defense and attack between pathogens and their hosts. It plays a critical role in the lytic, antimicrobial activity of human serum, the discovery of which was an early milestone in immunology, and decades of study have unraveled the interplay between proteins that lead to the formation of immune pores in microbial membranes. Assembly of the membrane attack complex (MAC) is the end product of a complex series of biochemical interactions in which initially soluble complement proteins bind and undergo dramatic structural rearrangements to form a transmembrane pore. To understand the molecular mechanism and kinetics underpinning how and when the MAC assembly becomes cytolytic, we sought to track the progression of the complement terminal pathway at the level of single pores. Using HS-PORT imaging on supported model membranes, we visualize the initial interactions of complement proteins with the membrane, and resolve the kinetics of MAC pore formation.
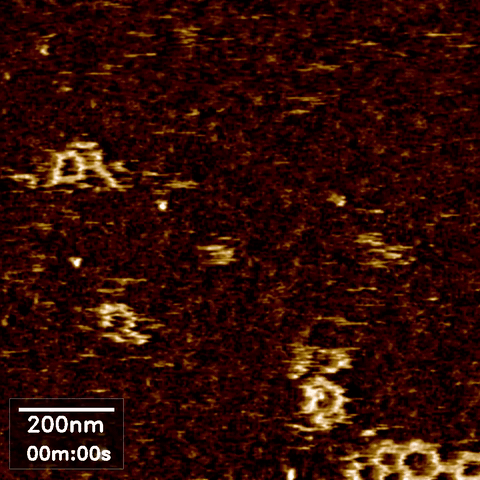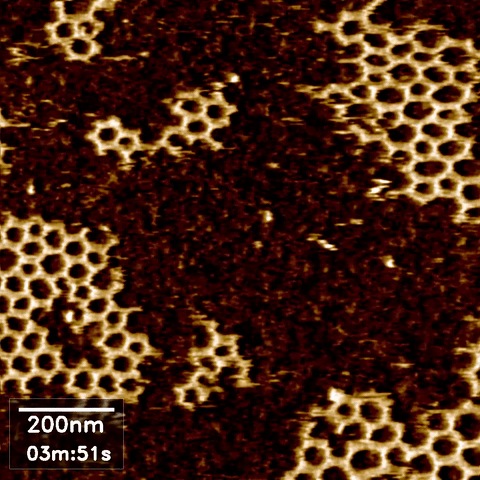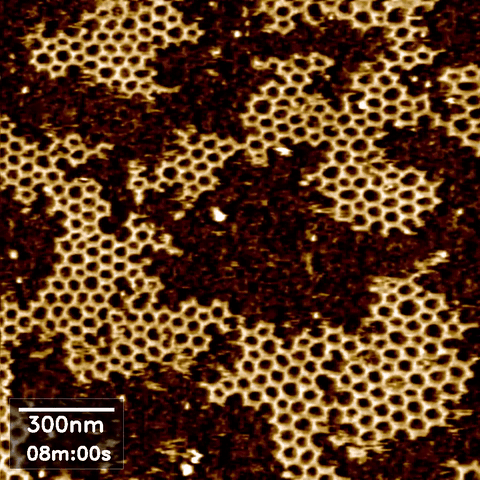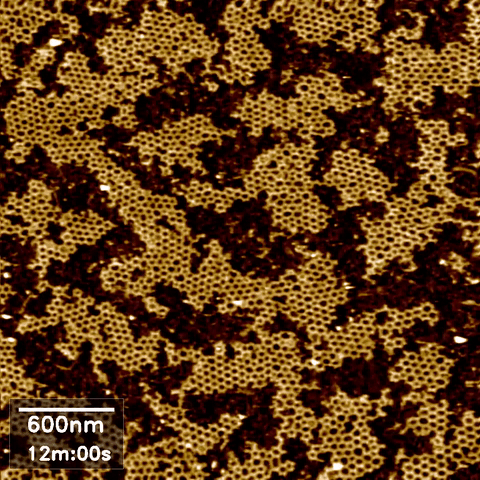
Over the last 35 years, DNA has been exploited to construct a wide range of nanostructures because of its excellent programming capability and structural simplicity. We utilized the capabilities of our new technique to resolve the self-assembly dynamics of hexagonal lattices formed using DNA nanotechnology.
Please note that the publication lists from Infoscience integrated into the EPFL website, lab or people pages are frozen following the launch of the new version of platform. The owners of these pages are invited to recreate their publication list from Infoscience. For any assistance, please consult the Infoscience help or contact support.
High-speed photothermal off-resonance atomic force microscopy reveals assembly routes of centriolar scaffold protein SAS-6
Nature Nanotechnology. 2018-08-13. Vol. 13, num. 8, p. 696-701. DOI : 10.1038/s41565-018-0149-4.Please note that the publication lists from Infoscience integrated into the EPFL website, lab or people pages are frozen following the launch of the new version of platform. The owners of these pages are invited to recreate their publication list from Infoscience. For any assistance, please consult the Infoscience help or contact support.
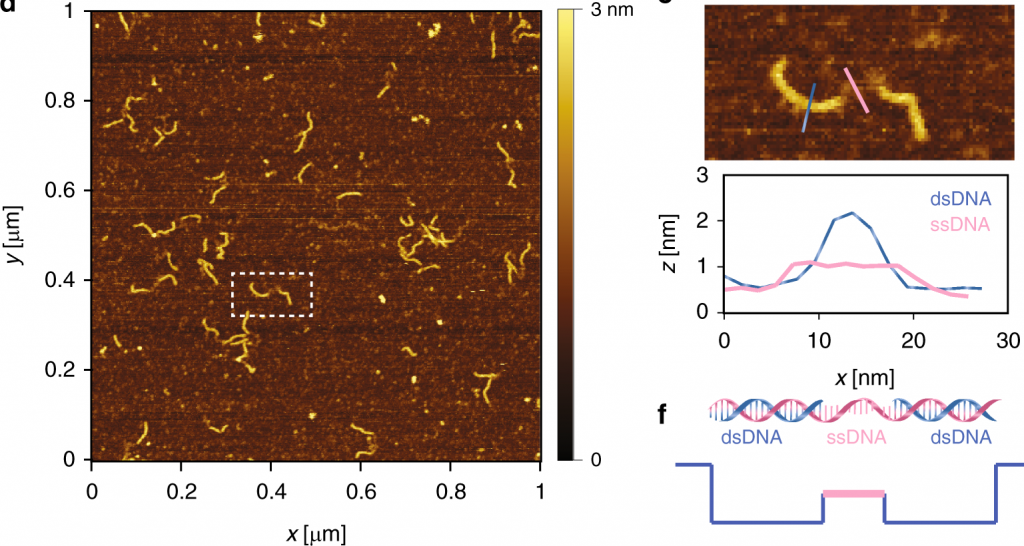
Detecting topological variations of DNA at single-molecule level
Nature Communications. 2019. Vol. 10, p. 3. DOI : 10.1038/s41467-018-07924-1.Please note that the publication lists from Infoscience integrated into the EPFL website, lab or people pages are frozen following the launch of the new version of platform. The owners of these pages are invited to recreate their publication list from Infoscience. For any assistance, please consult the Infoscience help or contact support.