Are you passionate about the nanoscale and its gateway, the AFM? Would you like to make use of your microfabrication skills to work on cutting-edge MEMS technology? Are you interested in observing biology at its smallest scale? Or do you want to push the imaging speeds of AFM through innovative control?
If any of those apply to you, then LBNI is the lab for your semester or master project!
Development of AFM cantilevers
Semester / Master project

Introduction
Two primary research goals in AFM are increasing the imaging speed and improving the ease of use to expand the potential range of applications. To address these objectives, we have started developing polymer based [1], self-sensing cantilevers [2] to improve the mechanical bandwidth of the cantilevers and be able to use a simple electrical readout to detect the deflection of the cantilever. These self-sensing devices are a promising replacement for the current state of the art cantilevers that are used with a complex laser based optical readout. You will find more information about our devices here.
Scope
While we are able to fabricate such cantilevers with high yield, we are now looking to improve them and adapt them for specific AFM applications. To help us do so, we are looking for highly motivated students who want to learn multidisciplinary skills. In joining us, you will be collaborating with a PhD student and you will get to know all about the fabrication of MEMS devices the EPFL cleanrooms, the CMi. In addition you will learn how to perform self-sensing AFM imaging. A successful completion will be subject to publication.
Interested? The different research axes are listed below!
Our current cantilevers have no tips and we are forced to add them one by one manually, which is very time consuming. To address this problem, you will be charged with developing a process to add tips onto our cantilevers directly during their fabrication.

In various AFM techniques, conductive tips are used to probe the surface. Since our cantilevers have embedded electronics, we experience cross-talk effects between them and the tip-connection. You will have to modify the process flow such as to add a grounded metal connection between the two signal lines, then use the fabricated cantilevers for electrical measurements in AFM.

We currently use the piezoresistive effect in poly-silicon to sense the deflection of our cantilevers. This material has a gauge factor of about 30. There are other materials, for instance single crystal silicon, that exhibit much higher gauge factors. You will be charged to modify the current process flow such as to replace the poly-silicon with single crystal silicon sensors.
What you will learn
- Cleanroom fabrication skills like photolithography, dry/wet etching, thin film deposition
- Creative thinking to find ways of manufacturing complex devices with standard microfabrication techniques
- Layout design for photolithography
- AFM imaging
Desired skills
- Fluency in English
- Autonomy
- Enjoys learning and facing new challenges
- Theoretical knowledge of microfabrication techniques (ex: fundamentals of microfabrication, materials and technology of microfabrication, MEMS practicals)
References
[1] Adams, Jonathan D., et al. “Harnessing the damping properties of materials for high-speed atomic force microscopy.” Nature nanotechnology (2015).
[2] Fantner, G. E., Adams, J. D. & HOSSEINI, N. Multilayer mems cantilevers. (2018).
Contact
At LBNI, we develop instruments at the forefront of nanoscale biology and one of our key strengths is instrumentation for atomic force microscopy (AFM). We develop high-speed AFM to push the current limits of imaging speeds. We believe that open hardware is the way forward to share the instruments and technologies that we develop, to the wider scientific community.
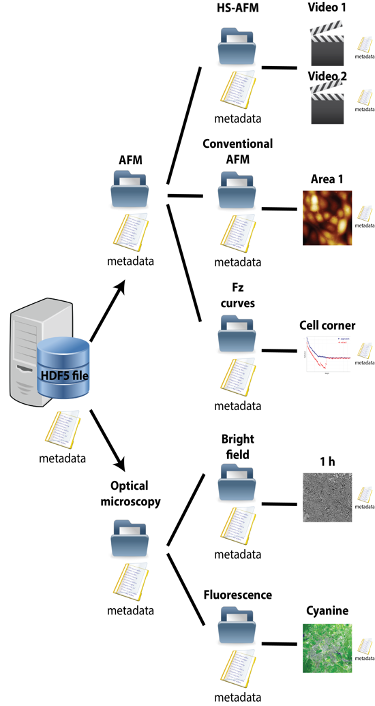
Open-source data schemes are becoming increasingly important for scientific data storage. They enable sharing of software, data, and knowledge at early phases in the scientific process, as well as proper long-term preservation storage. In this project, we aim to implement the HDF5 and USID open standards on an already well-established ORD worldwide project at LBNI: the Open-Source SPM controller [1]. Its current file scheme presents several weaknesses(maximum file size, metadata attachment, long term data curation, etc.) that we intend to overcome with those modern standards. Thus, the implementation of this scheme will enable the access to a lively open software ecosystem called Pycroscopy, where the Open-Source SPM project will take advantage of all the functionalities and tools already existing on that ecosystem based on the HDF5 and USID open standards.
In the frame of this project, we will need to design the different data structures represented via the USID model for the different techniques involved on the Open-Source SPM controller (Fig. 1): AFM, spectroscopy- AFM, HS-AFM, SICM, SEM, AFSEM, SNOM, optical microscopy and fluorescence. We will define the adequate USID structure to fulfil the different data generated by the previously enumerated techniques. At this point, it is crucial to design a proper metadata template for each technique, that can contain all the relevant information. After, all the USID designed schemes that will generate the HDF5 files from the acquired data on the Open-Source SPM controller will be coded using C/C++ language, generating Dynamic-link libraries (DLLs), which will be directly embedded on the LabView code that controls the Open-Source SPM controller.
What you will learn:
- AFM imaging and image processing.
- Interface with open-source SPM software control.
- Knowledge of the modern and open HDF5-USID scheme for advanced data storage.
Desired skills:
- Fluency in English.
- C/C++ programming experience, LabView knowledge is a plus.
- Ability and willingness to pick up new skills independently.
[1] https://www.epfl.ch/labs/lbni/spm-controller-software/
Contact
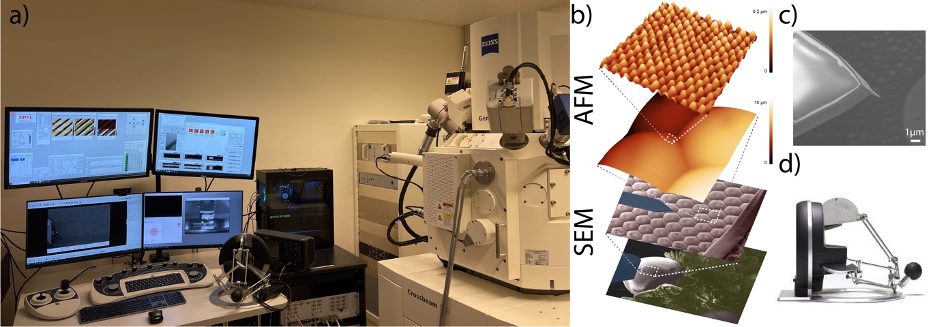
Our laboratory (LBNI) has a wealth of experience and expertise developing a wide range of instruments at the forefront of nanoscale biology. One of our key strengths is instrumentation for scanning probe microscopy (SPM) techniques such as atomic force microscopy (AFM). In SPM instruments, the controller plays a crucial role as servers as a control hub for all the sensors and actuators used in the system. In our lab, we have designed and developed a modular SPM controller (hardware and software) that provides open access at all levels, delivering at the same time research-grade performance in an open hardware philosophy, with several users around the world [1]. This gives the flexibility to adapt the controller to any setup and needs, making it easier to add new functionalities or devices. Thus, this controller has been deployed for various SPM research projects, including high-speed AFM, scanning electron microcopy (SEM), and other novel techniques. Recently, we have implemented an AFM inside of a CrossBeam 550L FIB/SEM system (AFSEM) (Fig 1a), where we combined the sub-nanometer resolution of an AFM with a wide range area and fast scan imaging of the SEM, enabling the imaging of challenging samples (Fig 1b).
We aim to use the nanometer size cantilever tip as a nano-manipulator tool (Fig 1c), controlled by an Omega.x haptic device (Fig 1d), programing in LabView the required routines to control the cantilever XYZ position. To achieve this, it is vital to optimize the communication architecture to reach fast controller/haptic device update rate (> 1 kHz), and to design a well-integrated human interface to facilitate its usability. This device will also serve as a tool for the functional understanding of AFM.
What you will learn:
- AFM and SEM imaging and image processing.
- Interface with open-source SPM software control.
- The principles of nano-manipulation based on Omega.x haptic devices.
Desired skills:
- Fluency in English.
- LabView programming experience.
- Ability and willingness to pick up new skills independently.
[1] https://www.epfl.ch/labs/lbni/spm-controller-software/
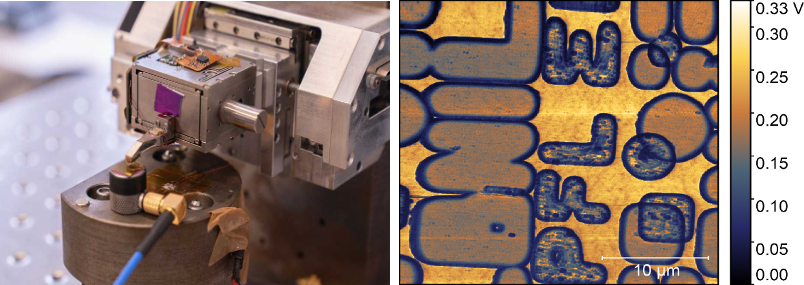
In the age of correlative microscopy, combining two modalities of microscopy techniques is fascinating. However, such combinations are to be facilitated to take the most advantage of this combination. Combining an atomic force microscope (AFM) and a scanning electron microscope (SEM) (Crossbeam 550L FIB/SEM system), allows us to have sub-nanometer resolution of an AFM with fast scan imaging of the SEM in challenging samples. However, beyond imaging AFM can help us achieve contrast based on different mechanical characteristics of materials which can be combined with the chemical contrast achieved by SEM’s backscattered detectors.
Contact resonance (CR) measurements are a very unique of set of surface and subsurface imaging where the tip-sample contact is ultrasonically excited at a specified frequency and the amplitude and phase of the contact resonance is monitored. This contact resonance being sensitive to the sample’s local mechanical properties, CR-AFM can help us map the various mechanical properties of the sample. Varying the force (setpoint) for the contact mode imaging, this method can be used to perform sub-surface imaging which can complement the SEM’s in depth backscattered analysis.
We aim to facilitate these CR-AFM measurements in our AFM which operates inside the SEM chamber. To achieve this, we need to facilitate sending a signal to the sample inside the SEM and use frequency tracking with a lock-in amplifier to discern the different material present below the surface and complement it with SEM imaging.
What you will learn:
- AFM (CR-AFM) imaging and SEM imaging
- Interpret material contrast based on frequency tracking
- Use of Lock-in amplifier and instrumentation for vacuum technology
Desired skills:
- Fluency in English
- Ability and willingness to learn new skills independently
Contact
Laboratory LBNI and LBEN have developed system for nanopore-based label-free single-molecule sensor – scanning ion conduction spectroscopy (SICS)1, Fig 1A. It utilizes a glass nanopore mounted on a three-dimensional nano positioner to spatially select molecules tethered on a glass surface for controlled translocations. This technique grants us the ability to actively choose specific regions of interest on the DNA and scan molecules a predetermined number of times and at a controlled velocity, Fig 1B.
LBNI group has a wealth of experience and expertise developing a wide range of instruments at the forefront of nanoscale biology. One of our key strengths is instrumentation for scanning probe microscopy (SPM) techniques such as scanning ion conduction microscopy (SICM). In SPM instruments, the controller plays a crucial role as servers as a control hub for all the sensors and actuators used in the system. In our lab, we have designed and developed a modular SPM controller (hardware and software) that provides open access at all levels, delivering at the same time research-grade performance in an open hardware philosophy, with several users around the world2. This gives the flexibility to adapt the controller to any setup and needs, making it easier to add new functionalities or devices. Thus, this controller has been deployed for various SPM research projects, including high-speed AFM, scanning electron microcopy (SEM), and other novel techniques.
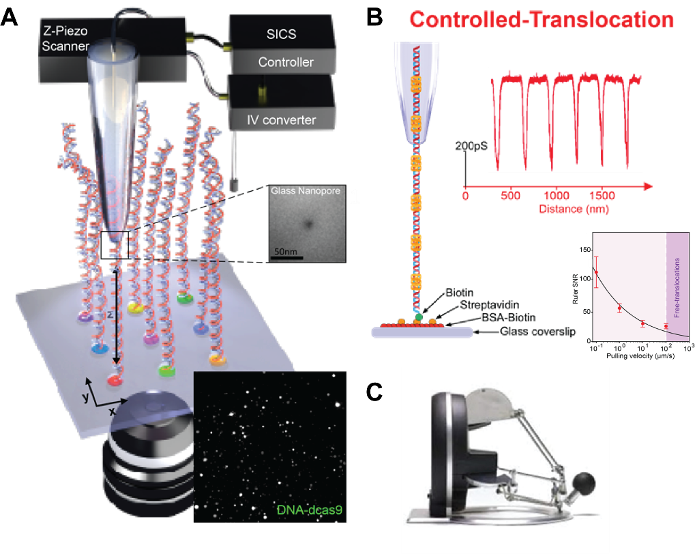
We are offering a semester/master project or an internship opportunity to work on implementing use of Omega.x haptic device (Fig 1c) for sensing DNA analytes with nanometer-sized capillary. Manipulation in XYZ, with the right routines showing how close the tip is to the surface, would be programmed in LabView software. To achieve this, it is vital to optimize the communication architecture to reach fast controller/haptic device update rate (> 1 kHz), and to design a well-integrated human interface to facilitate its usability.
What you will learn
- SICS sensing and data processing
- Interface with open-source SPM software control.
- The principles of nano-manipulation based on Omega.x haptic devices.
Desired skills
- Fluency in English.
- LabView programming experience.
- Ability and willingness to pick up new skills independently.
Reference:
- Leitao, S. M. et al. Spatially multiplexed single-molecule translocations through a nanopore at controlled speeds. Nat. Nanotechnol. 18, 1078–1084 (2023).
- https://www.epfl.ch/labs/lbni/spm-controller-software/
Introduction
Atomic force microscopy (AFM) can take high-resolution images of nanoscale objects in various environments. Since conventional AFM imaging is too slow to capture dynamic processes of, for example, behavior and interaction of biomolecules, high speed AFM (HS-AFM) has been developed by optimizing the software and hardware in the instrument for fast scanning.
AFM images often require some post-processing in an open source software called Gwyddion. Data obtained from AFM imaging software Nanoscope can be directly imported, processed to flatten, remove lines and scars, and define the zero-level and contrast, then exported as a common image format such as jpeg. However, our existing software packages were not designed to handle the volume of data produced during HS AFM imaging, where we obtain hundreds of images per experimental run.
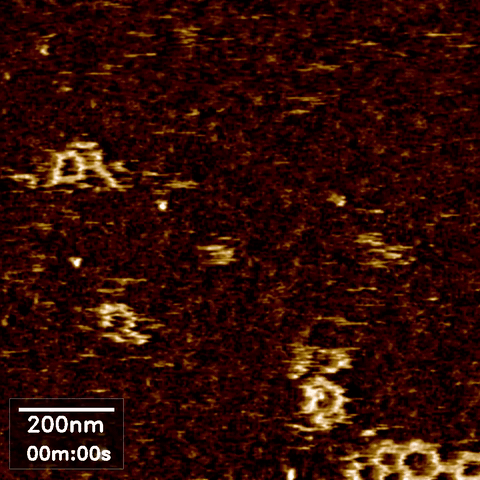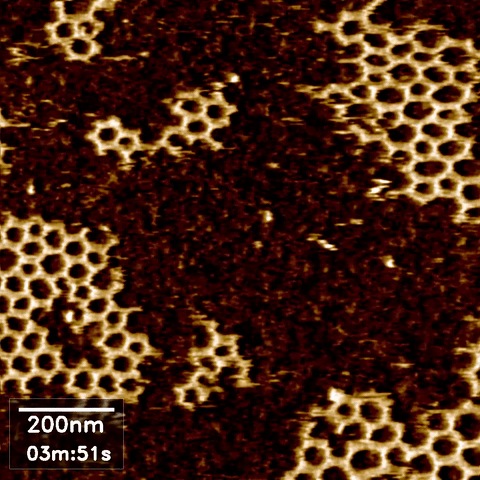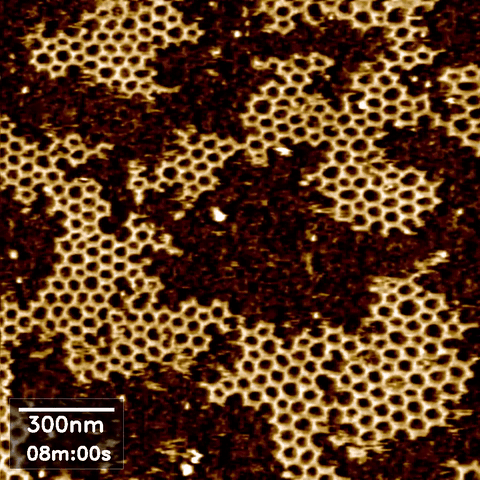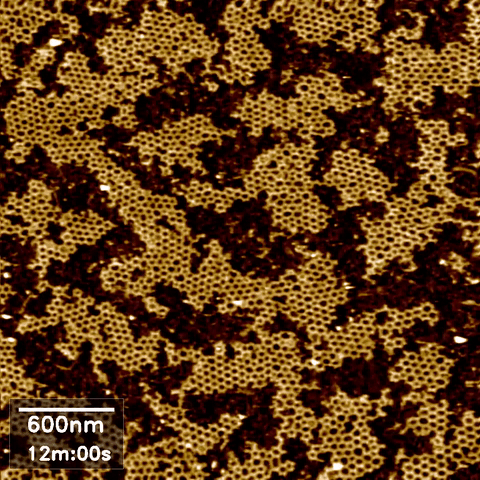
Scope
In high speed AFM imaging, we obtain a large quantity of images, which would take a significant amount of time to process with Gwyddion one by one. We are therefore looking for an interested Masters or semester project student to develop more efficient or automated AFM image processing for converting a series of HS-AFM images from the same experiment to a video format.
Project Task
Your task will be to design and test and a plugin for Gwyddion to efficiently process HS AFM data into videos.
What you will learn
- AFM imaging
- AFM image processing
- Interface with open source AFM image processing software
Desired skills
- Fluency in English
- Coding basics (Python)
- Ability and willingness to pick up new skills independently
References
Please note that the publication lists from Infoscience integrated into the EPFL website, lab or people pages are frozen following the launch of the new version of platform. The owners of these pages are invited to recreate their publication list from Infoscience. For any assistance, please consult the Infoscience help or contact support.
Large-scale analysis of high-speed atomic force microscopy data sets using adaptive image processing
Beilstein Journal of Nanotechnology. 2012. Vol. 3, p. 747-758. DOI : 10.3762/bjnano.3.84.Please note that the publication lists from Infoscience integrated into the EPFL website, lab or people pages are frozen following the launch of the new version of platform. The owners of these pages are invited to recreate their publication list from Infoscience. For any assistance, please consult the Infoscience help or contact support.



