Electron Microscopes can now provide the nanometer resolution that is needed to image synapses, and therefore connections, while Light Microscopes see at the micrometer resolution required to model the 3D structure of the dendritic network. Since both the arborescence and the connections are integral parts of the brain’s wiring diagram, combining these two modalities is critically important. In fact, these microscopes now routinely produce high-resolution imagery in such large quantities that the bottleneck becomes automated processing and interpretation, which is needed for such data to be exploited to its full potential.
Neuronal structures acquired using a Light Microscope look radically different when imaged using an Electron Microscope. Since the graph geometrical and topological structure may be the only property shared across modalities, graph matching becomes the only effective registration means. We will therefore use our Computer Vision expertise to propose a new approach for matching graph structures, which can handle these cases while being robust to topological differences between the two graphs and even changes in the distances between vertices. We will test our algorithms in aligning blood vessels or nerve fibers, acquired at different times, scales, viewpoints, or using different modalities, which may result in vastly diverse image appearances.
E. Serradell, P. Glowacki, J. Kybic, F. Moreno-Noguer, P. Fua
Algorithm
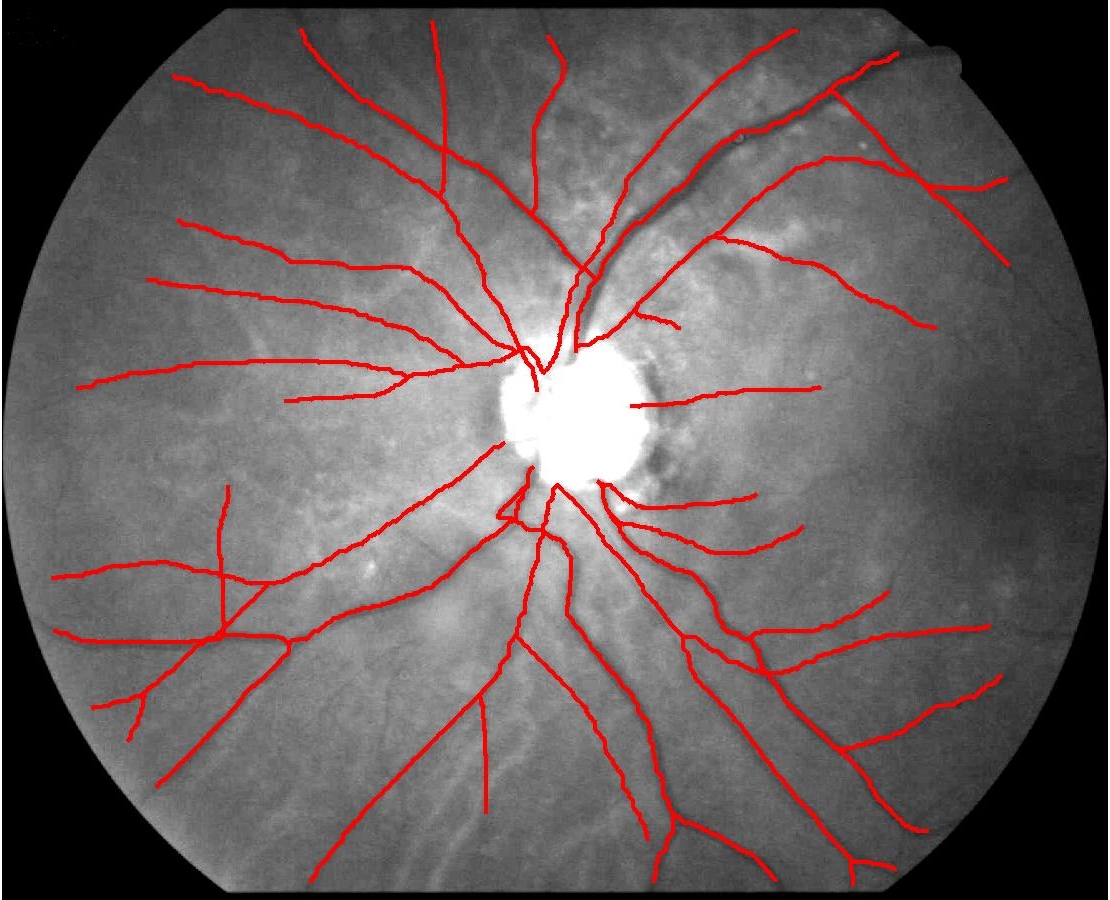
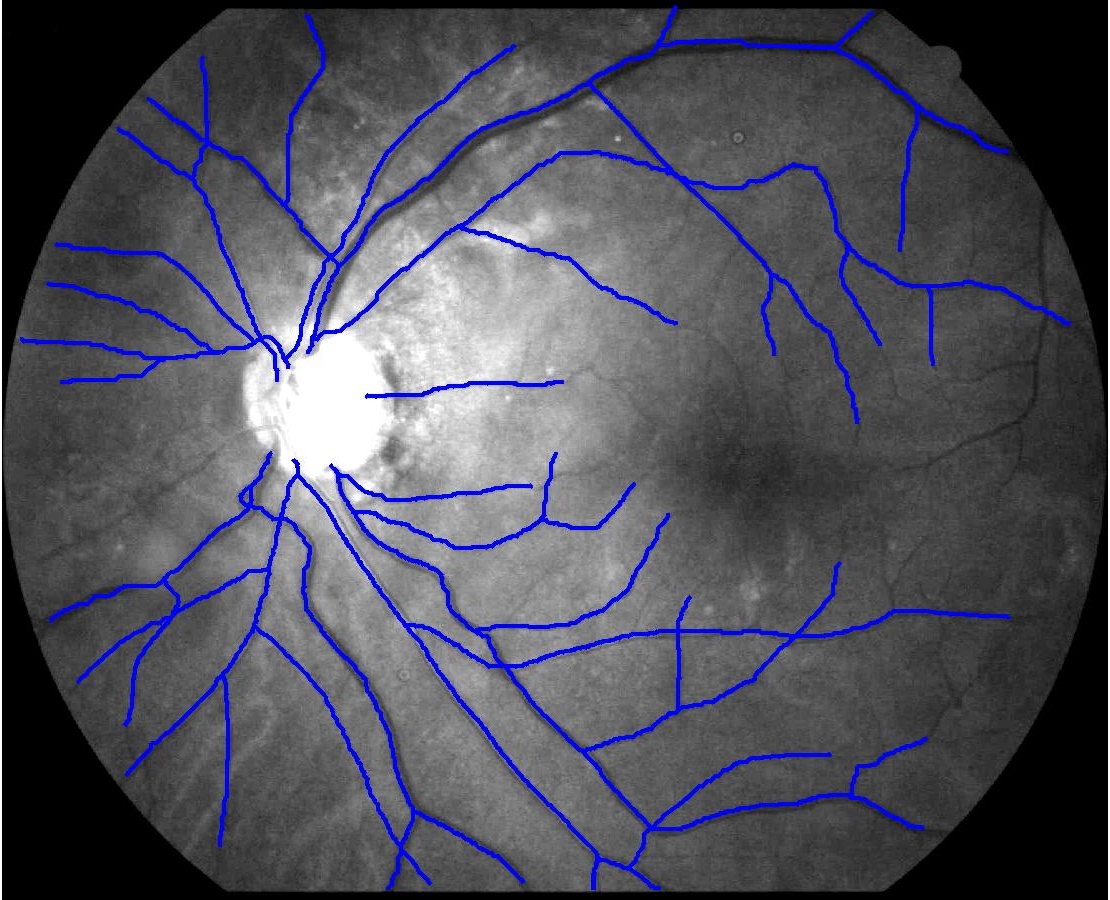
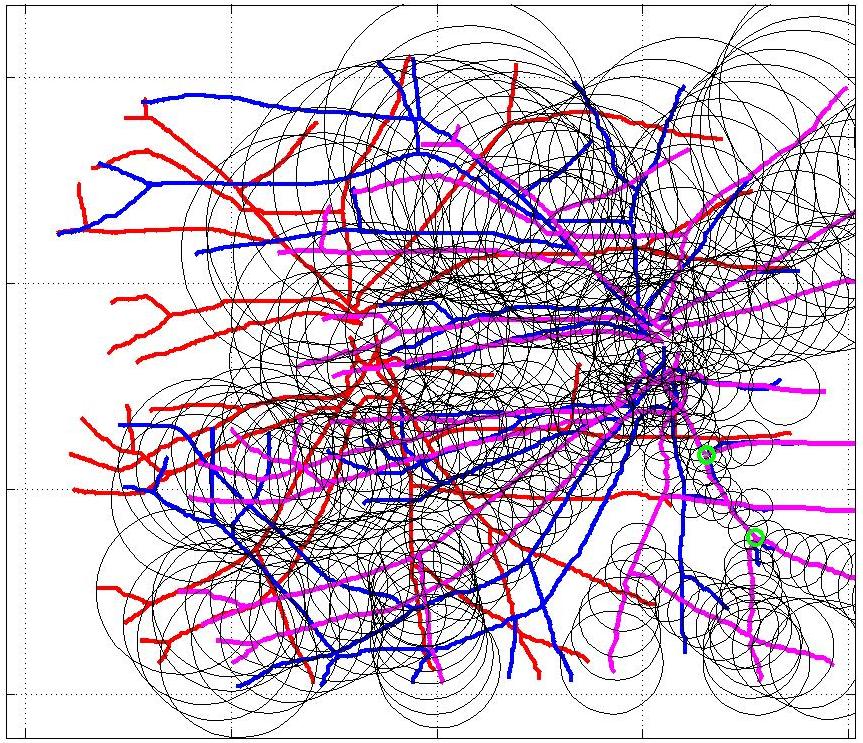
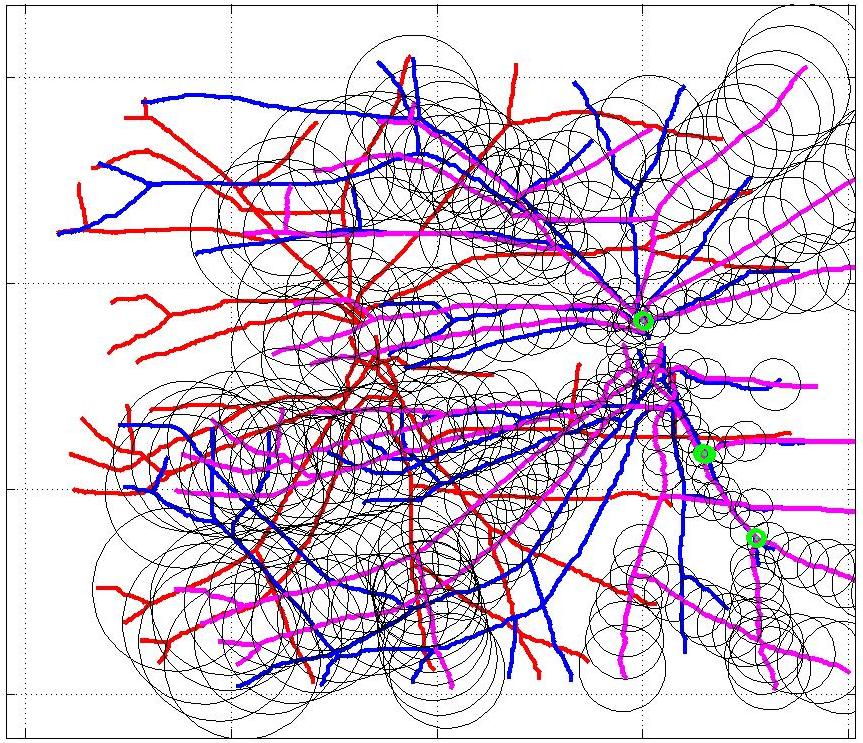
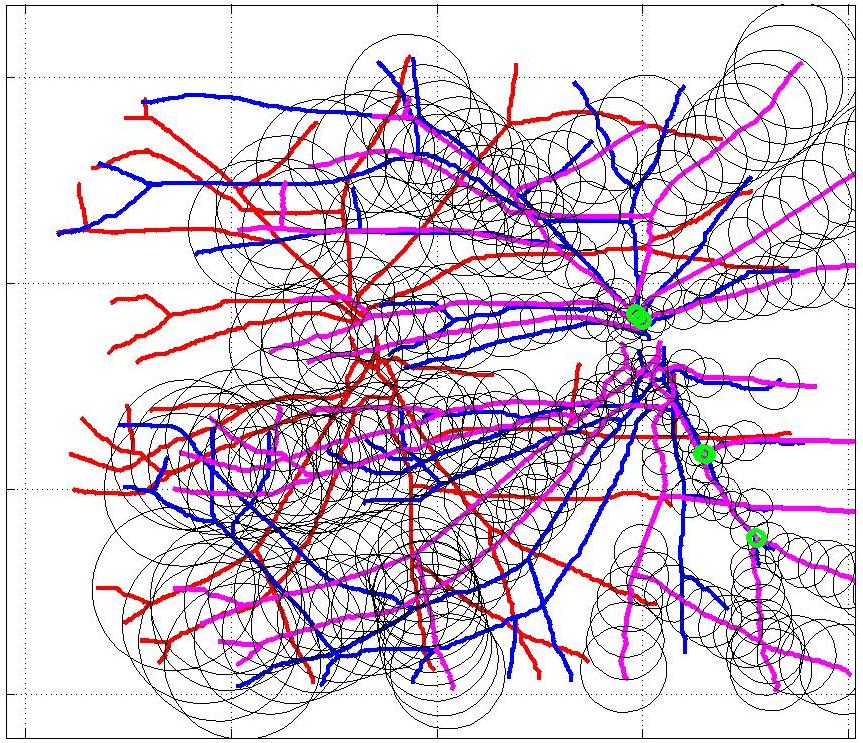
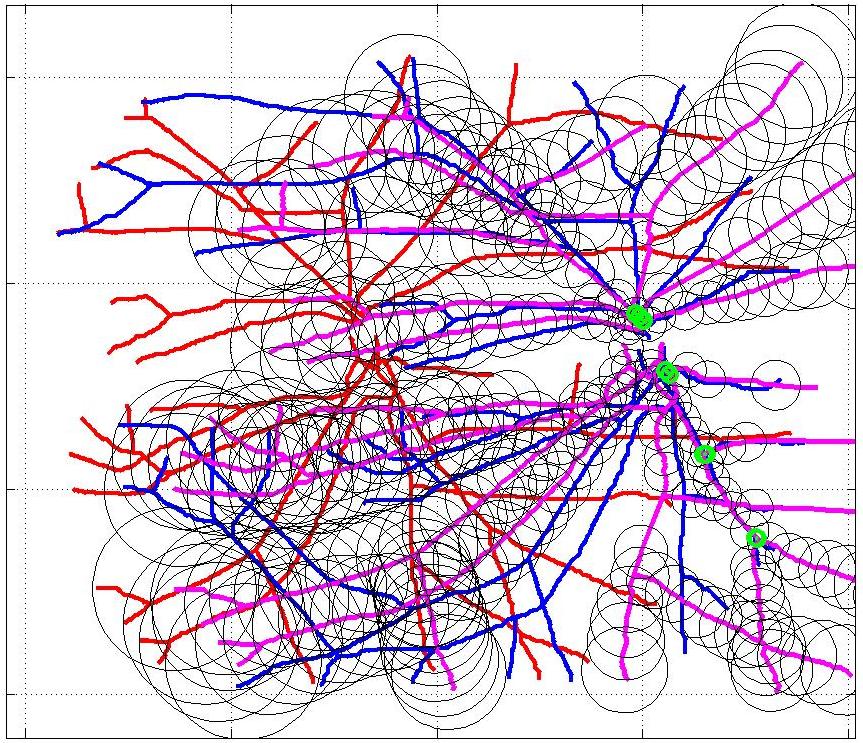
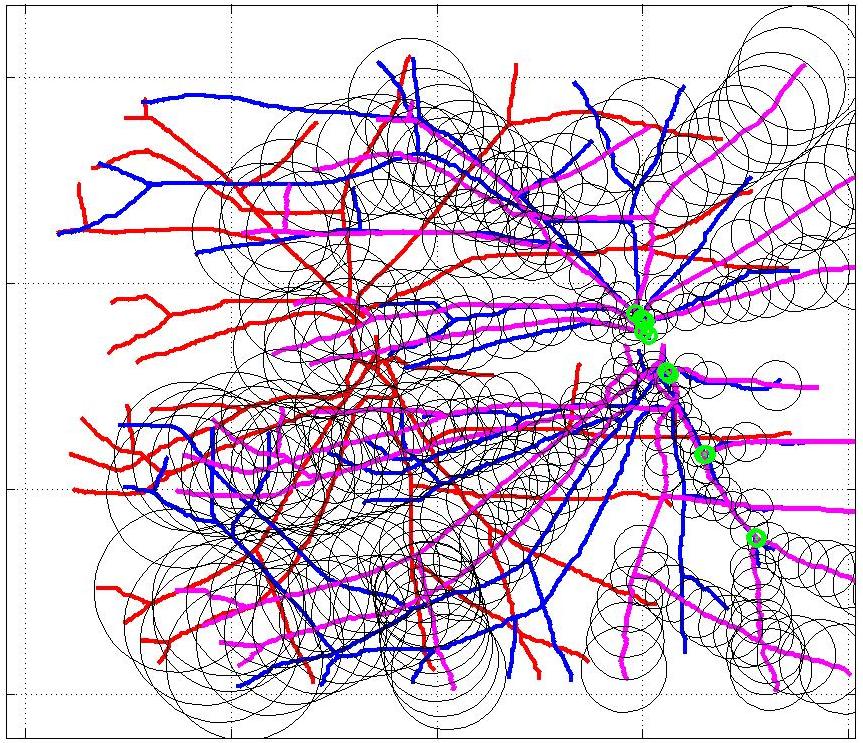
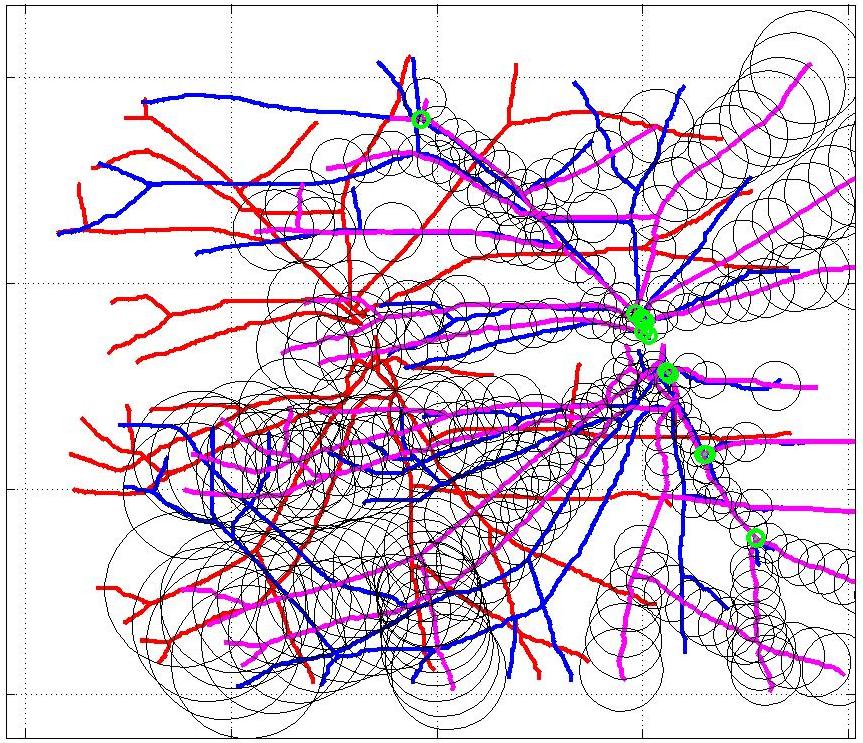
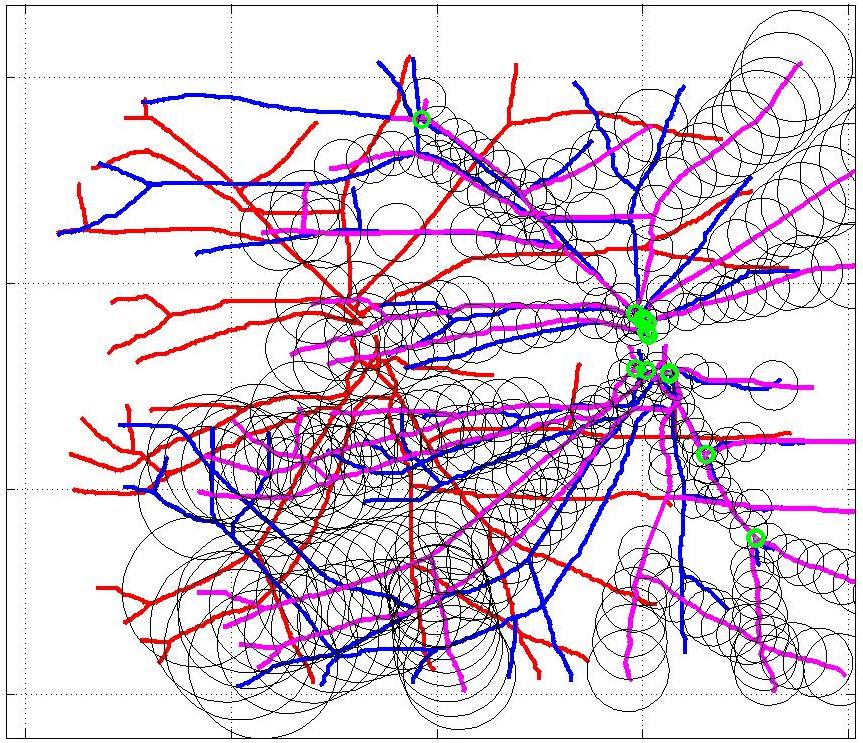
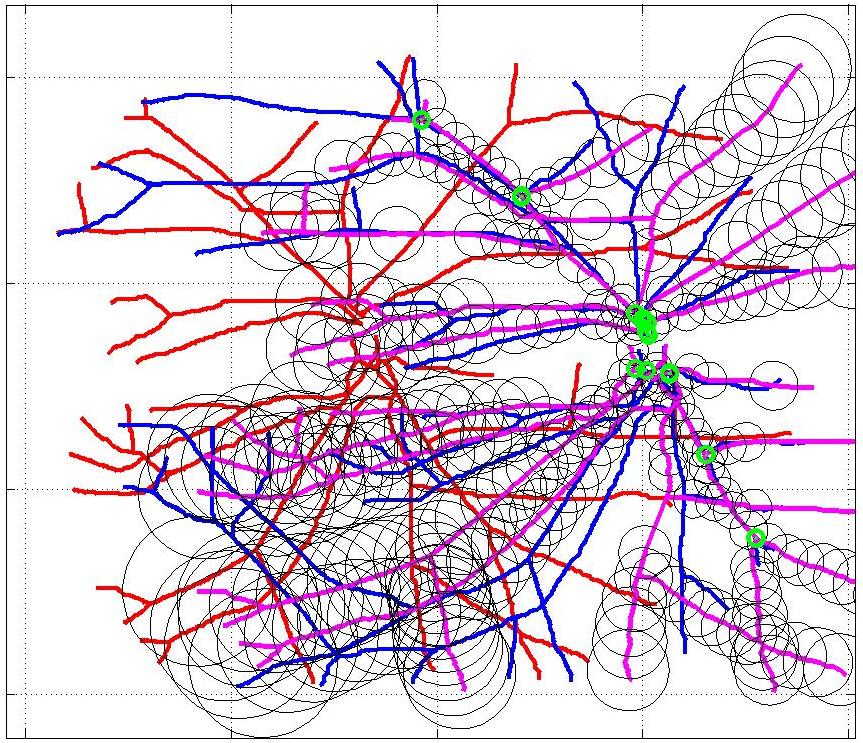
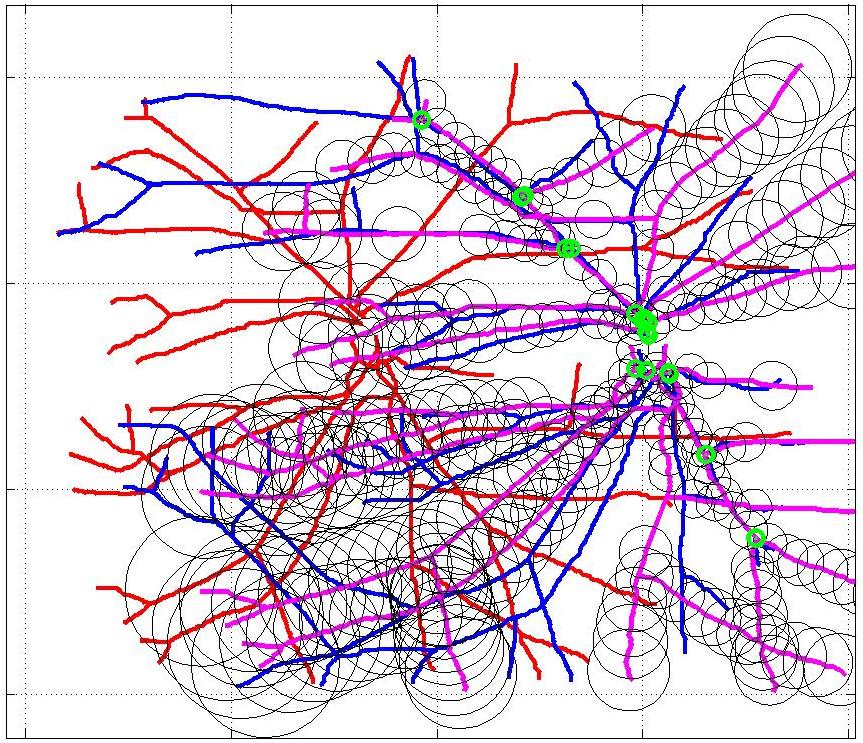
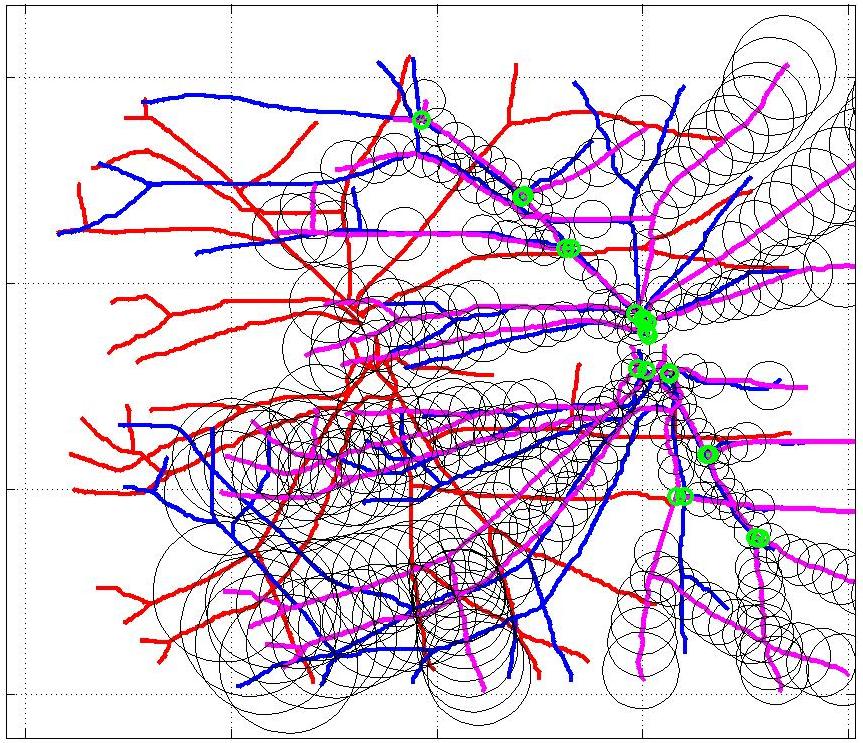
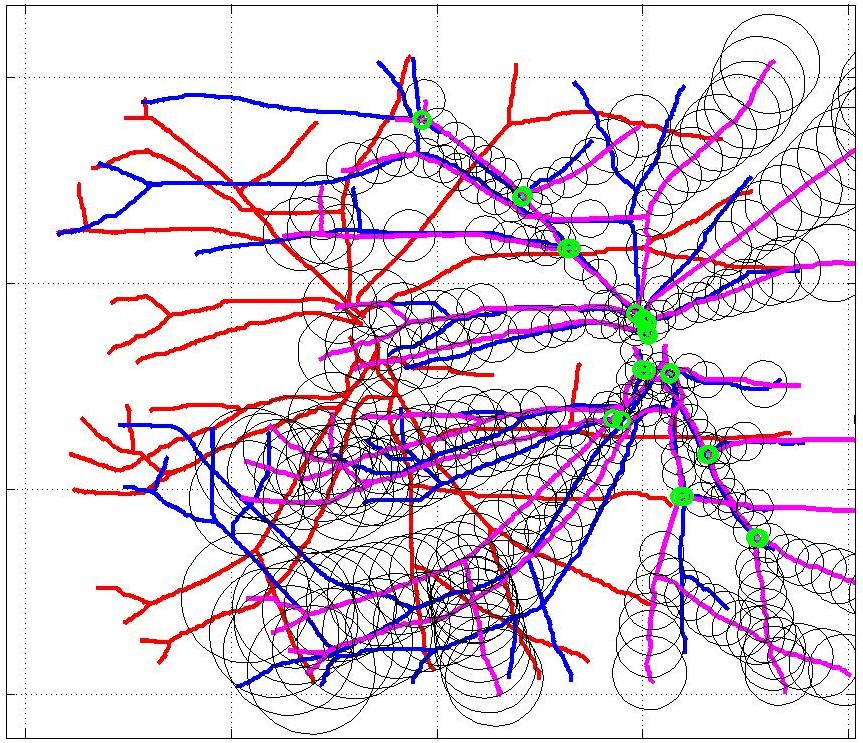
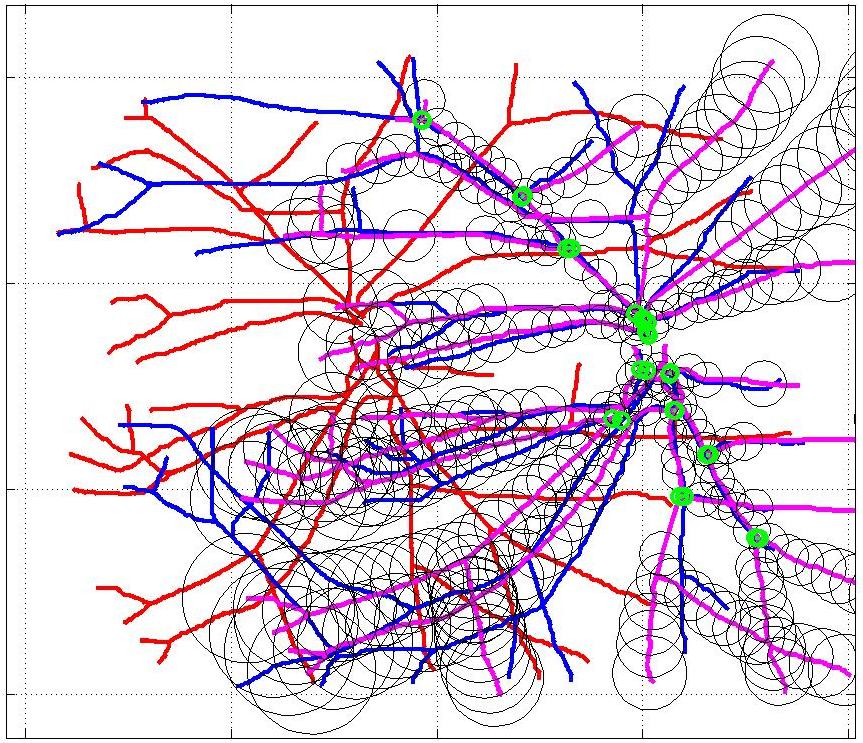
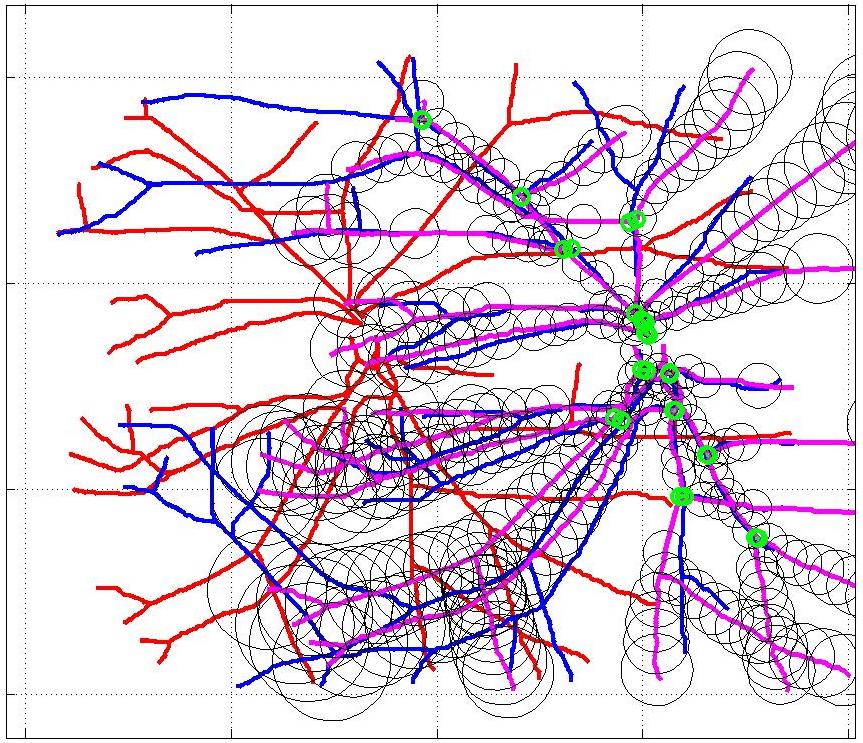
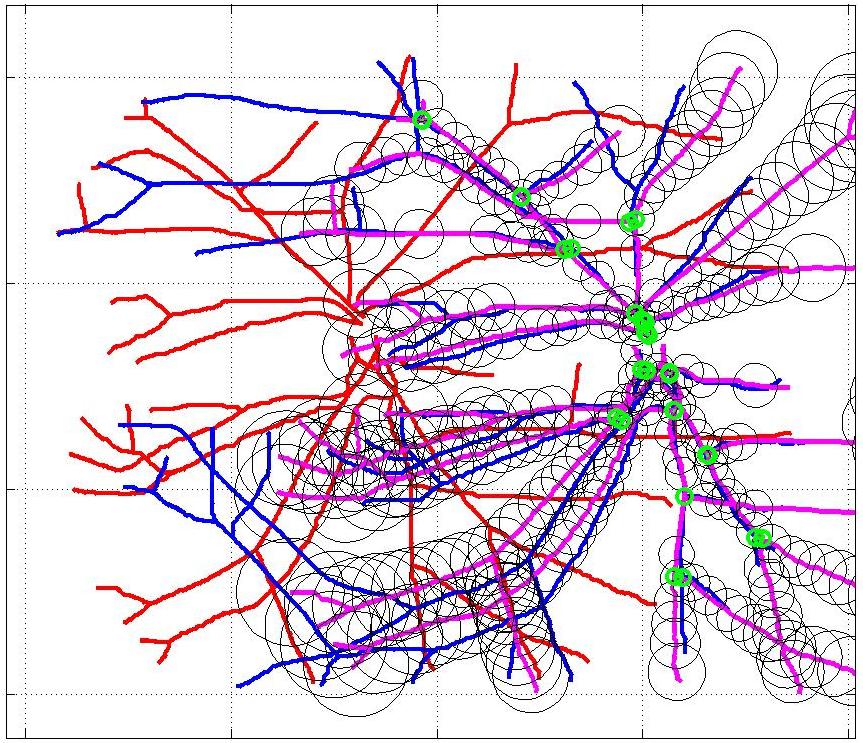
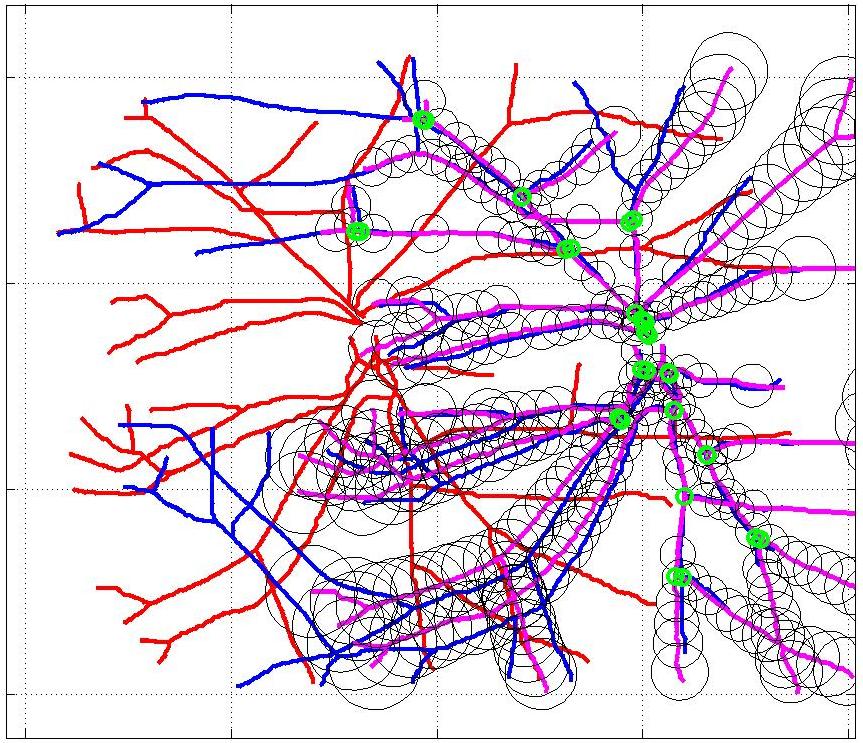
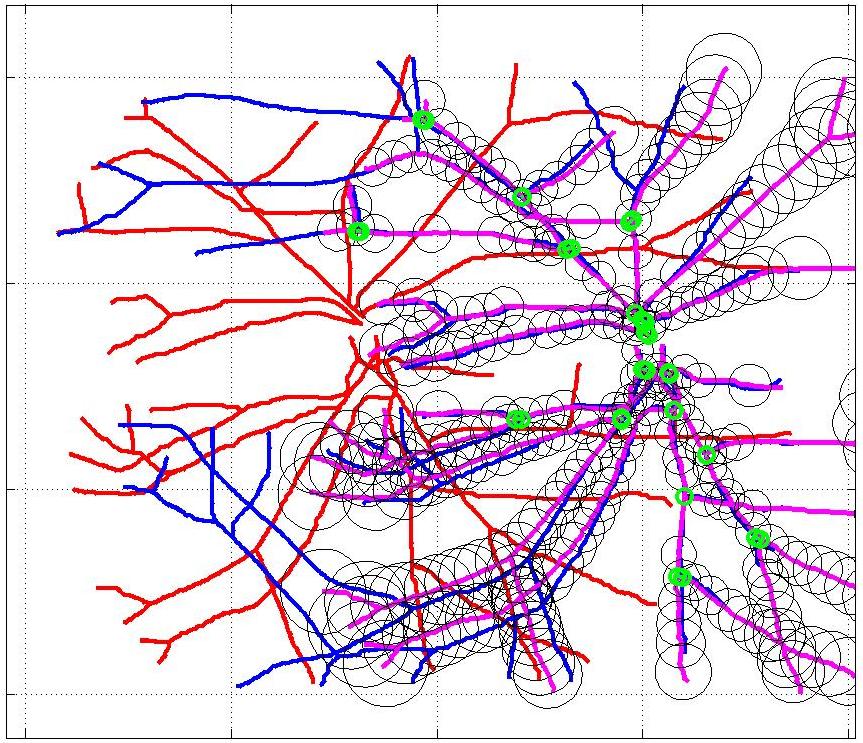
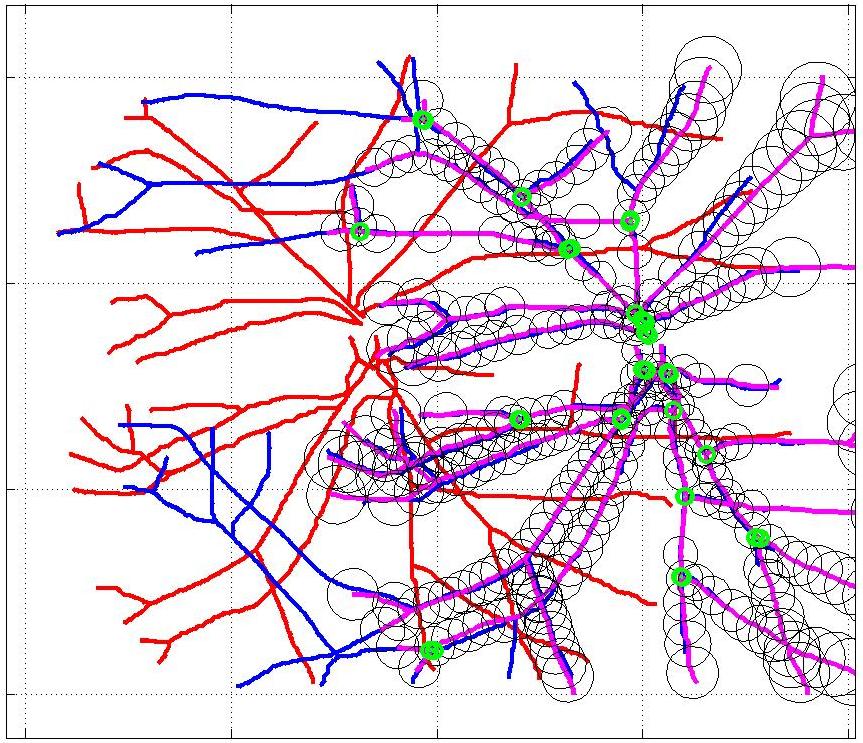
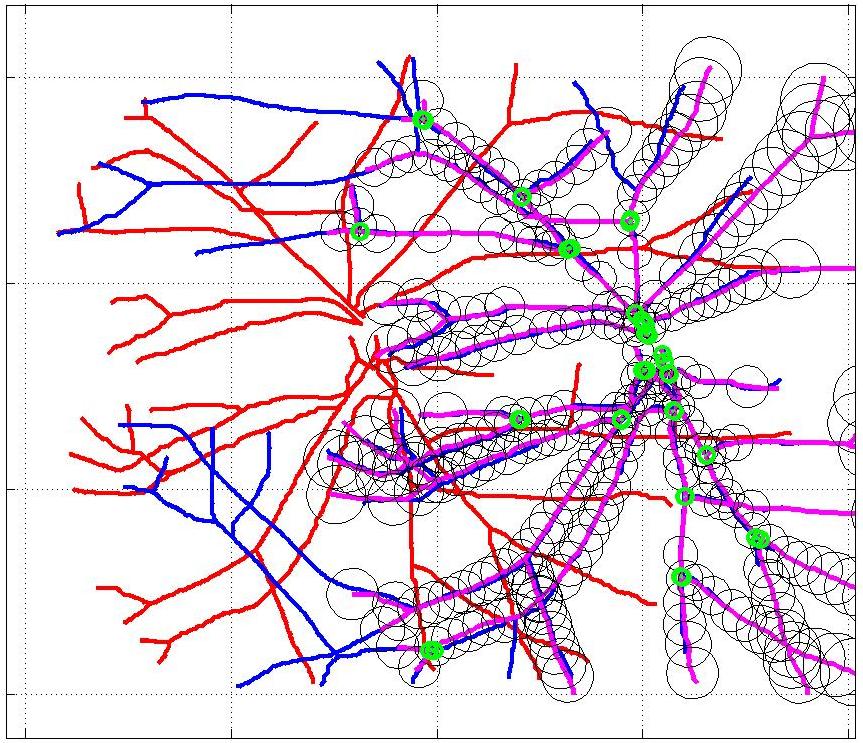
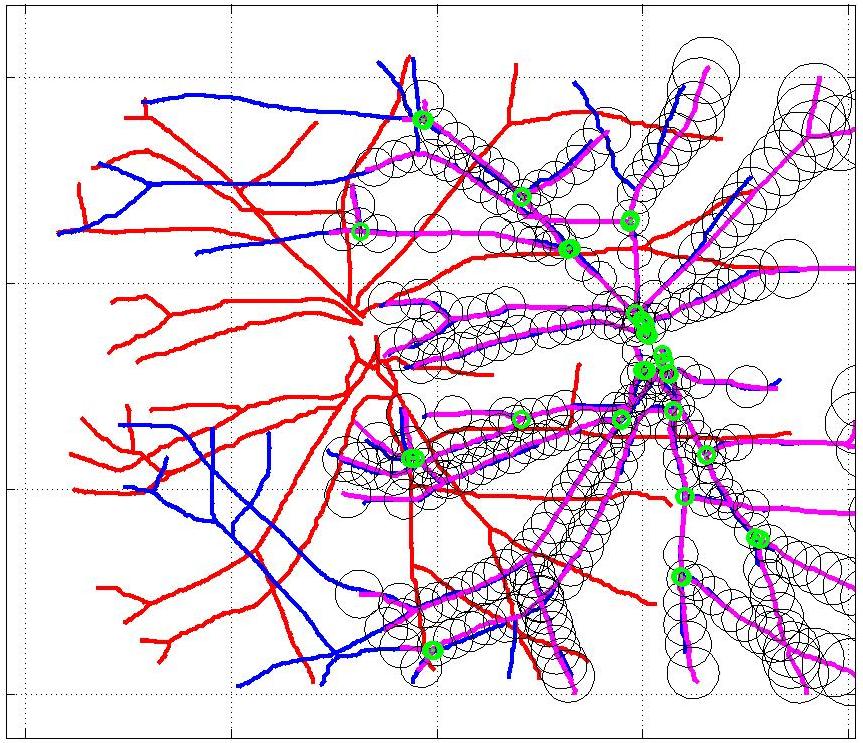
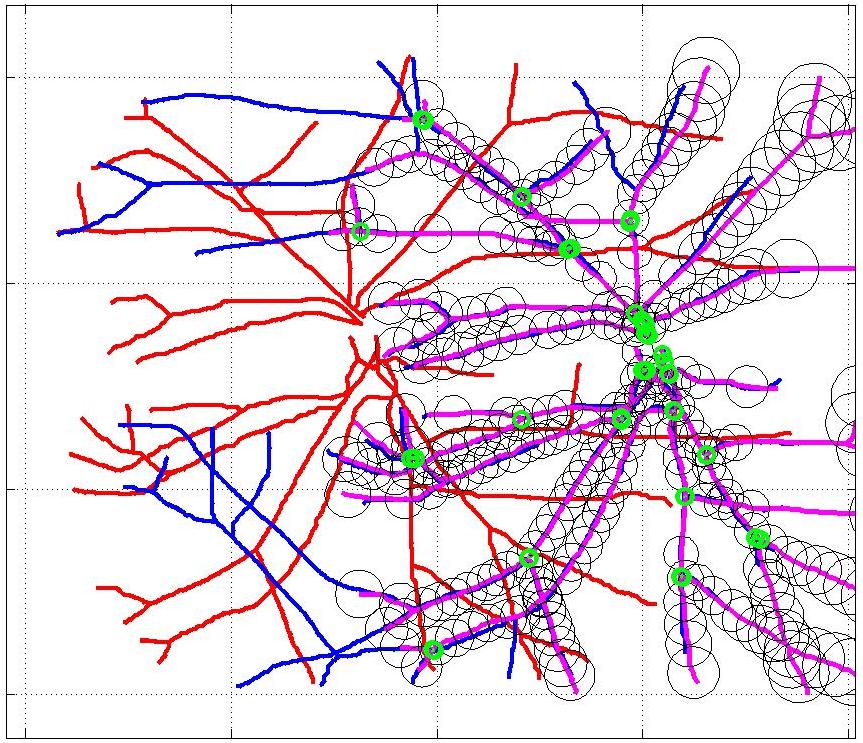
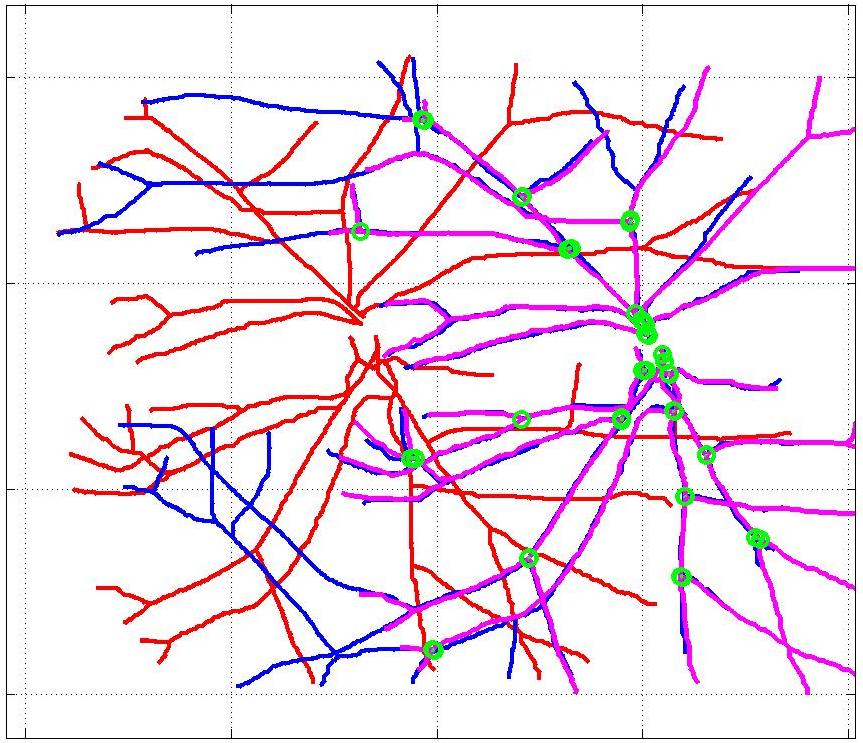
This animation walks through the intermediate steps of our algorithm to show how the predictions of the geometric mapping from one data set to the other, modelled as a Gaussian Process, are progressively refined as more correspondences are added.
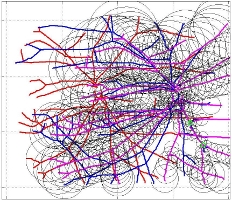
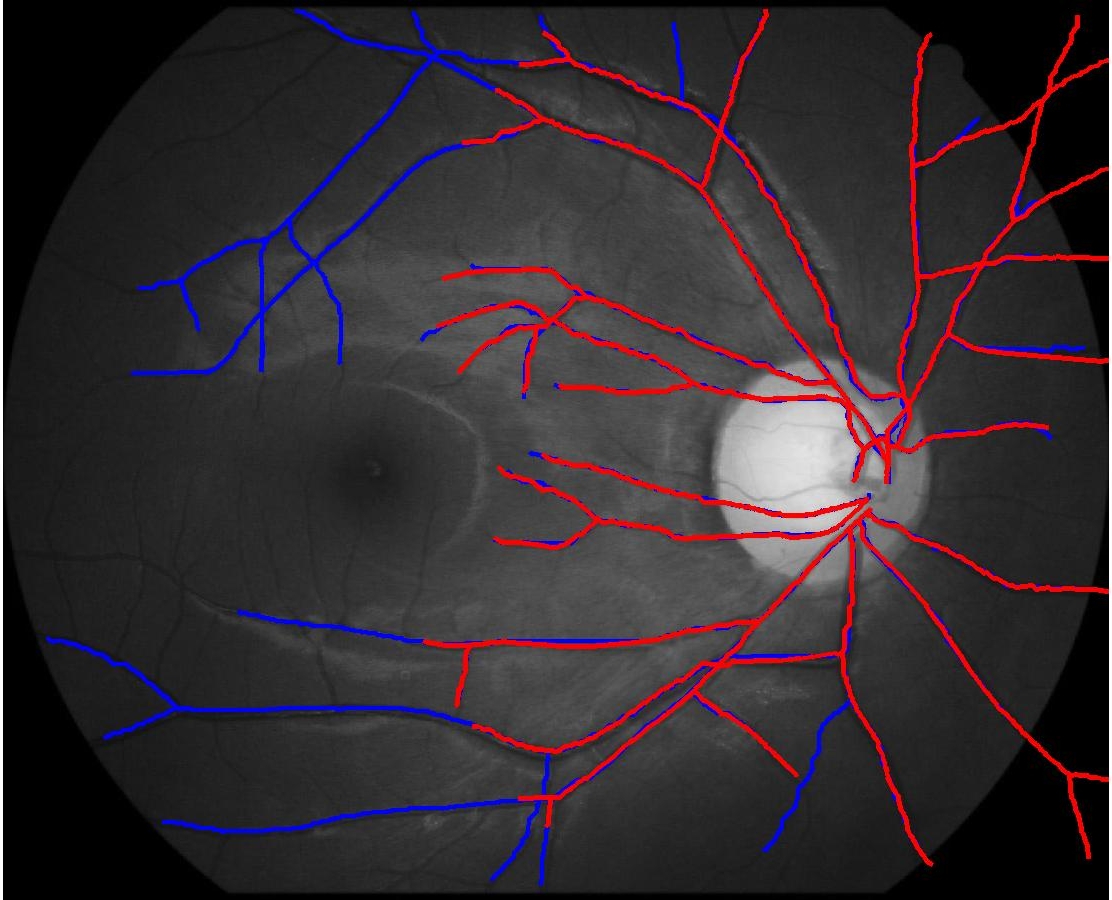
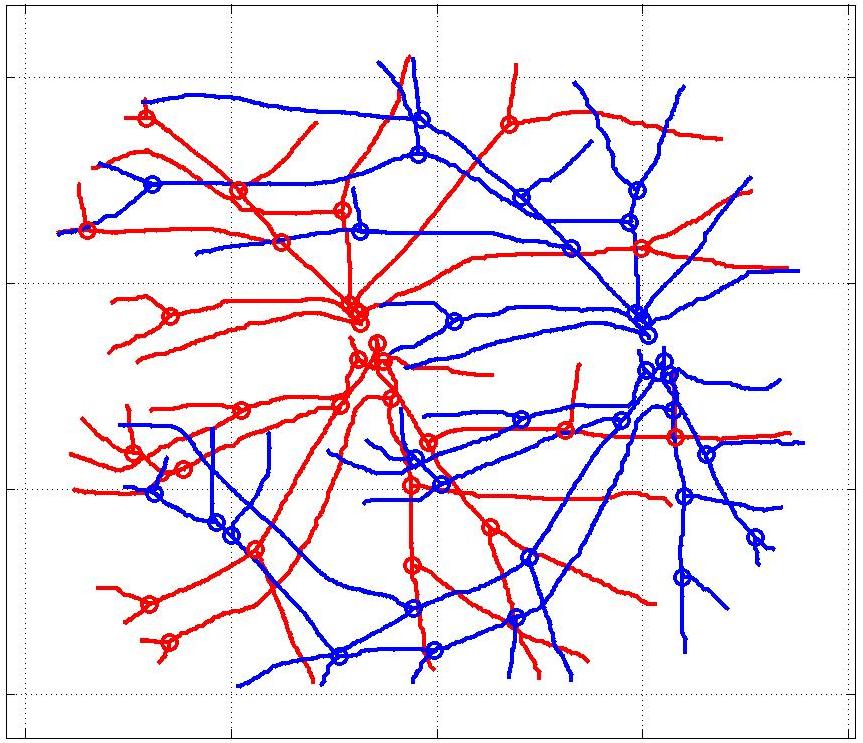
In this case, the graphs to match are embedded in 2D and represent vessel segmentations from retinal scans. Click on the prev/next button to advance the slideshow.
Results
Neuronal stacks
Embed of video is only possible from Mediaspace, SwitchTube, Vimeo or Youtube
Registration of the 3D neuronal stacks extracted from the brain tissue using two different modalities: LM and EM. The common neuron structures are used to align the two image stacks. First, we obtain a coarse alignment of the neuronal bifurcations. Then, we refine it using the whole structure and obtain a geometric mapping which can be applied to every voxel of the image stack.
Blood vessel alignment
Embed of video is only possible from Mediaspace, SwitchTube, Vimeo or Youtube
Embed of video is only possible from Mediaspace, SwitchTube, Vimeo or Youtube
The retinal fundus vascular graphs are deformed from one image to the next because the camera is looking from different points of view. This results in apparent distortions of the curved retinal surface’s projection.
In the sequence of 2D X-ray angiography images of a beating heart, the deformations suffered by the blood vessels are much more non-linear. We align the vessel structures segmented on two different moments of the heart cycle.
Materials
You can find the Matlab source code in the Software section.
References
Robust Non-Rigid Registration of 2D and 3D Graphs
2012. Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, June 2012. p. 996-1003. DOI : 10.1109/CVPR.2012.6247776.Non-Rigid Graph Registration using Active Testing Search
IEEE Transactions on Pattern Analysis and Machine Intelligence. 2015. Vol. 37, num. 3, p. 625-638. DOI : 10.1109/TPAMI.2014.2343235.Contacts
| Eduard Serradell | [e-mail] |
| Przemisław Głowacki | [e-mail] |
| Jan Kybic | [e-mail] |
| Francesc Moreno-Noguer | [e-mail] |
| Pascal Fua | [e-mail] |