In the current medical context, 3D medical imaging is integral to enhancing disease diagnosis and facilitating effective treatment planning. However, conventional techniques for image analysis predominantly focus on voxel-based methodologies, which can often be laborious and prone to inaccuracies. Our objective is to utilize advanced deep learning methods to transcend this voxel-centric approach, thereby promoting a more efficient and precise interpretation of 3D medical images.
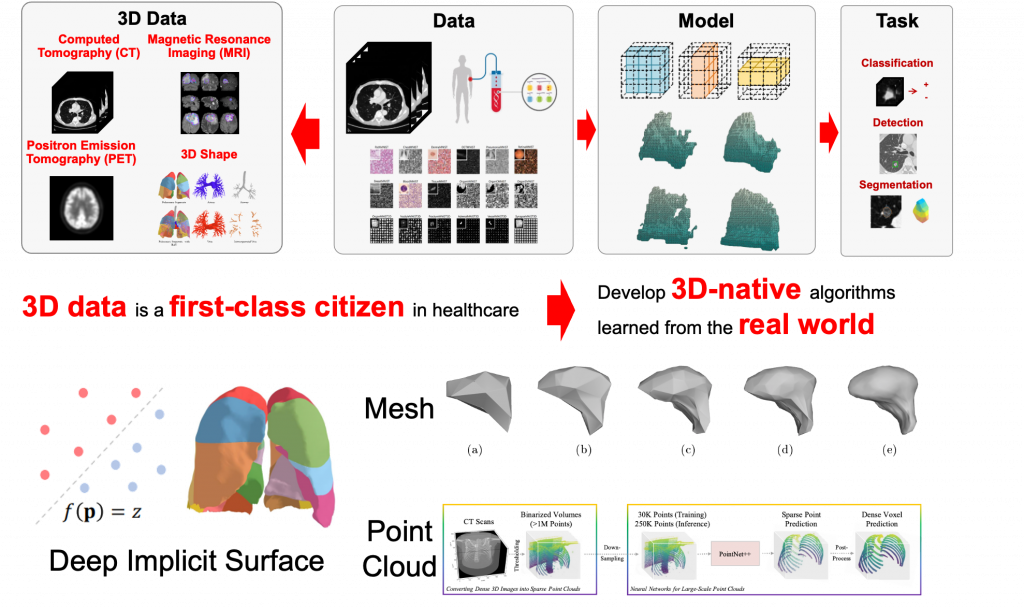
At CVLab, our research focus is divided into the following three core areas:
- Deep implicit surface [1,2]
- Surface [3] and tetrahedral [4] mesh
- Point cloud [5]
Our approach involves leveraging deep learning as a data-driven method to automatically interpret and analyze medical images. This not only reduces reliance on human intervention but also minimizes potential inaccuracies. Simultaneously, we are committed to tackling high-dimensional problems to amplify the capabilities of traditional image analysis techniques, thereby enabling them to address more complex issues. Engaging with these challenges will not only push the boundaries of academic research but also cultivate practical solutions to real-world problems in healthcare.
Students participating in this project will concentrate on one of the provided areas.
Prerequisites:
- Proficiency in Python programming language
- Familiarity with deep learning and the PyTorch library
- Knowledge of 3D vision or computer graphics would be advantageous
- A passion for modeling real-world problems and resolving healthcare challenges!
Contact:
Interested students are requested to send an email to Jiancheng Yang ([email protected]).
Reference:
[1] Yang, J., Wickramasinghe, U., Ni, B. and Fua, P. “ImplicitAtlas: Learning deformable shape templates in medical imaging”. CVPR 2022.
[2] Yang, J., Shi, R., Wickramasinghe, U., Zhu, Q., Ni, B. and Fua, P. “Neural annotation refinement: Development of a new 3D dataset for adrenal gland analysis”. MICCAI 2022.
[3] Wickramasinghe, U., Jensen, P., Shah, M., Yang, J. and Fua, P. “Weakly Supervised Volumetric Image Segmentation with Deformed Templates”. MICCAI 2022.
[4] Gao, et al. “Learning deformable tetrahedral meshes for 3D reconstruction.” NeurIPS 2020.
[5] Yang, J., Gu, S., Wei, D., Pfister, H. and Ni, B. “RibSeg dataset and strong point cloud baselines for rib segmentation from CT scans”. MICCAI 2021.