Dr. Julian C Shillcock
The phase separation of intrinsically-disordered proteins into liquid droplets, or biomolecular condensates, has emerged as a major organisational principle of cells. Proteins within droplets interact in complex ways, and over time undergo aberrant phase transitions into misfolded states that lead to the growth of rigid fibrils. These fibrils are associated with devastating diseases including Alzheimer’s, Parkinson’s and Huntington’s disease. I use molecular simulations and biophysical modelling to explore the phase separation of Intrinsically-Disordered Proteins (IDP) into fluid droplets and the subsequent emergence of rigid fibrils.
Biological droplets are hundreds of nanometres to microns in size, which is beyond the scale accessible with atomistic Molecular dynamics.
I use coarse-grained simulations to study the liquid phase separation of model proteins (see image), and compare with atomistic Molecular Dynamics simulations of single proteins to link molecular details of IDPs to their phase separation. The droplet here is ~ 50 nm in diameter.

blank
Fibril formation takes place over hours to days, which is too slow even for coarse-grained simulation techniques like Dissipative Particle Dynamics; I am collaborating with Prof. John Ipsen of the University of Southern Denmark to develop thermodynamic models of protein aggregation and amyloid formation. These models will be compared with the coarse-grained simulations (e.g., DPD and Monte Carlo) to explore equilibrium and non-equilibrium models of fibril formation.
blank
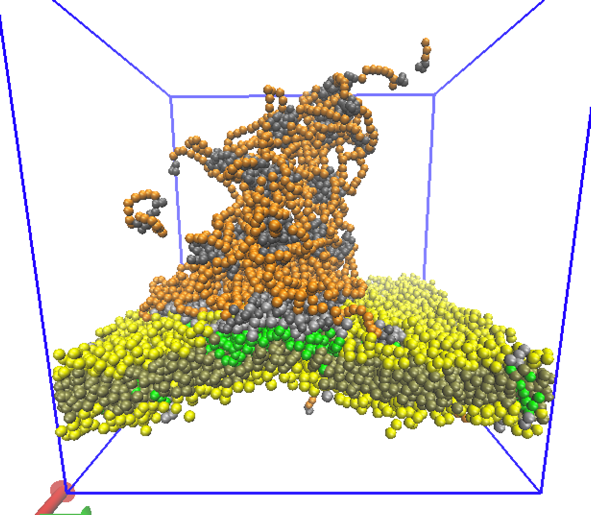
Finally, the formation of amyloid fibrils is associated with membrane interactions of IDPs, so I am developing models to explore how phase separating proteins interact with lipid membranes, and how binding may drive domain formation.
Blank
Challenges
1) How much molecular detail is required to simulate IDPs to an accuracy sufficient to predict LLPS and aggregation?
2) What experimental tools are needed to calibrate the theoretical and simulation models?
3) Can simulations span the dynamical range from the atomistic scale of single proteins to the micron-scale, slow dynamics of fibrilisation?
4) Can we accelerate molecular-scale simulations to follow LLPS on experimental system sizes of microns?
Blank
LMNN/Blue Brain Project Collaborations
We are exploring collaborations between LMNN and the Blue Brain Project in two areas:
1) Synaptic plasticity in health and disease
The Postsynaptic density (PSD) is an assembly of proteins, receptors and signalling molecules associated with the postsynaptic membrane of excitatory neurons that is involved with synaptic plasticity.
It has been proposed that the PSD forms by a process of liquid-liquid phase separation of the constituent intrinsically-disordered proteins. Key PSD proteins have been reconstituted into an in vitro PSD by Zeng et al. (Cell 174:1172, 2018). Neurological diseases result when this phase separation fails.
The goal of this project will be to identify how the physical chemical properties of the PSD depend on its constituents and to use this knowledge to increase the biological fidelity of synapse modelling in the BBP cortical simulations.
2) Topological quantification of phase separated droplets
Biomolecular condensates are porous droplets of disordered proteins. The proteins form reversible bonds that create a three-dimensional network or web. In disease conditions, these networks transform into a more rigid state. Newly-applied topological measures may be useful in quantifying the structural properties of condensates and identifying signatures of the healthy fluid phase and pathological rigid state.
Teaching
What I cannot build, I don’t understand.
I teach a Master’s course in Computational Cell Biology (BIOENG-455) in which students construct numerical models and run computer simulations of biological structures and processes, and compare the results quantitatively against experimental or simulation data.
blank
Open Positions
Coming soon