SEMESTER PROJECT PROPOSALS
1. Domain adaptation for ultrasound image quality enhancement with deep learning
This project focuses on improving the image quality of ultrasound images using a deep-learning-based image reconstruction techniques. The goal is to adapt the current CNN to a different setting and equipment configuration, enabling imaging at greater depths and wider fields of view. Various approaches, including transfer learning and training from scratch, will be explored using a new dataset. The project will be applied to fetal and liver imaging and 3D tracked systems.
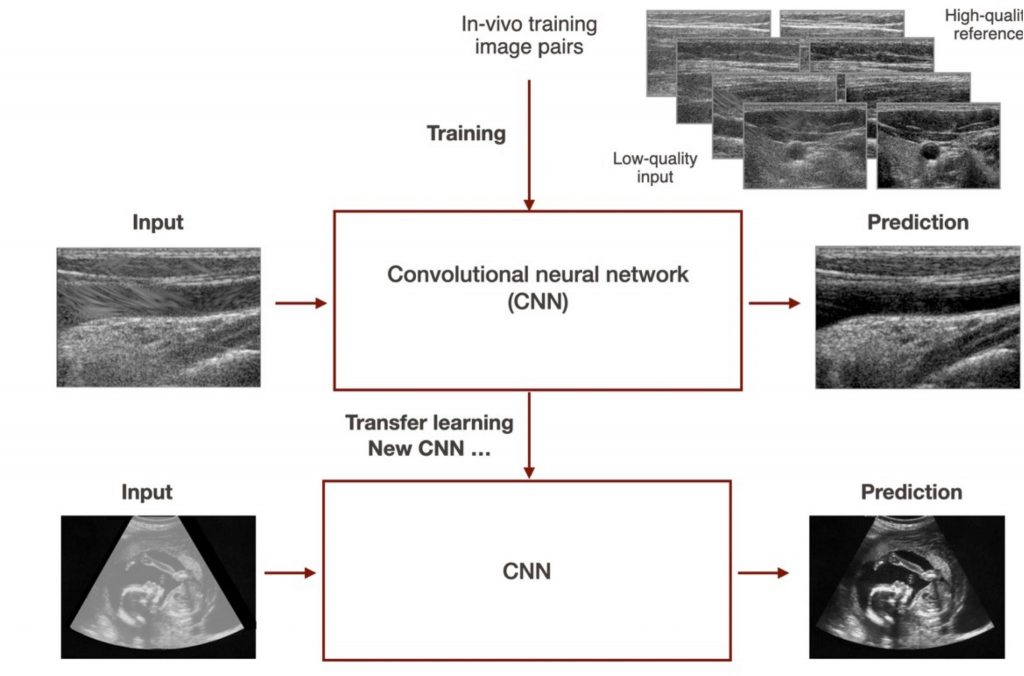
To acquire the dataset we will ask the student to use an ultrasound device after proper training, potentially receiving compensation for creating a large in vivo dataset.
Requirements: Signal processing and deep learning. Fluent in python and pytorch.
Assistants: Roser Viñals Terres ([email protected]) and Sandra Marcadent ([email protected])
Supervisor: Prof. Jean-Philippe Thiran
2. Deep learning for foetal head segmentation on 3D-tracked ultrasound images
Due to its non-invasive nature, ultrasound imaging is a widely employed technique in the field of obstetrics and gynaecology. It relies on emitting ultrasound waves into tissues and receiving echoes produced at tissue interfaces.
By integrating ultrasonography with an optical tracking system, such as infrared cameras and a reflective marker on the ultrasound probe, 3D-tracked ultrasound imaging enables the continuous recording of the image plane’s position in 3D space. By employing this technique, we can reconstruct large volumes and achieve 4D visualization by fitting 3D models to the generated images and point clouds. In the frame of a larger project in obstetrics*, we aim to develop such visualization system for the foetal head during childbirth.
One crucial step towards this goal is the automatic and real-time segmentation of the foetal head on ultrasound images. The segmentation process would consist of two main parts. First, we need to extract a mask and biometric parameters from independent 2D images. Second, we need to accurately segment the fine contours of the skull from a set of 3D-tracked images to extract a 3D point cloud.
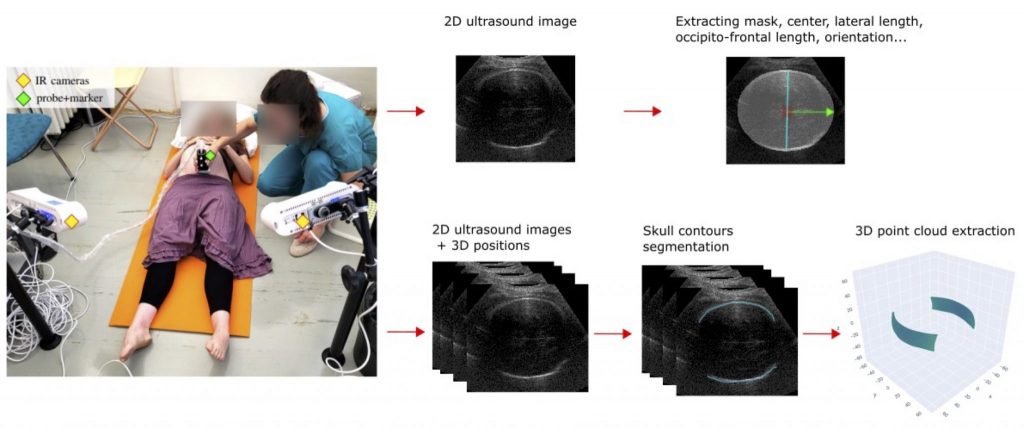
We are looking for a Master student to work on those two segmentation steps. The first part will involve the development and application of a deep learning approach, incorporating techniques such as transfer learning, data augmentation and the design of a task-specific training loss. For the second part, the student will develop a method utilizing the 3D spatial consistency amongst ultrasound images.
The ideal candidate for this project should have solid programming skills with proficiency in PyTorch and a strong foundation in image processing, machine learning and deep learning.
Assistants : Sandra Marcadent ([email protected]), Ayoub Tajja ([email protected])
Supervisor : Prof. Jean-Philippe Thiran
*You will find more information about it here: https://vimeo.com/321254812
3. Spherical Deconvolution Algorithms for Intra-Voxel Fiber Estimation and Brain Connectivity Mapping
The LTS5 Diffusion group focuses on brain tissue microstructure and structural connectivity –estimated by diffusion Magnetic Resonance Imaging (dMRI) data, with a particular focus on the reconstruction of the nerve fiber orientation distribution function (ODF) per voxel (see the figure below). This information is important for the reconstruction of the brain’s white matter by using fiber tracking algorithms (see ref [1]).
We have implemented various novel reconstruction algorithms (e.g., see refs. [2-5]) and we plan to develop a new generation of methods using Machine Learning techniques. The goals of this project are: (1) create a large database of fiber ODFs and corresponding dMRI signals, (2) Design, train, and optimize a neural network using this dataset, (3) predict the fiber ODFs from new dMRI data, and (4) compare the implemented algorithm with state-of-the-art techniques using both synthetic and real dMRI data acquired from human brains. The results will be published in international conferences and relevant journals.
Requirements: The project will be implemented in Python, so good knowledge is required. This project is ideal for a computer scientist, mathematician, physicist, or engineer interested in medical imaging, machine learning, signal processing, and optimization.
References:
[1] https://www.sciencedirect.com/science/article/abs/pii/S1053811914003541
[2] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4607500
[3] https://www.sciencedirect.com/science/article/abs/pii/S1053811918307699
[4] https://onlinelibrary.wiley.com/doi/10.1002/mrm.21917
Supervisors: Dr. Erick J. Canales-Rodríguez ([email protected]), Dr. Gabriel Girard ([email protected]), and Prof. Jean-Philippe Thiran
4. Myelin Water Imaging Using T2 Relaxometry
Myelin is a lipid-rich substance that surrounds the axons in the brain, which is essential for the proper functioning of the nervous system. Myelin water imaging is a magnetic resonance imaging (MRI) method that can be used to quantify and visualize myelination in the brain and spinal cord in vivo. The signal coming from the MRI machine (using a multi-echo T2 relaxometry sequence) can be decomposed into components, including that originated by water molecules trapped between the lipid bilayers of myelin. The correct estimation of this component provides a myelin-specific MRI biomarker to monitor brain changes in cerebral white matter. Myelin quantification has important implications for understanding various neurodegenerative diseases, including multiple sclerosis.
We are looking for a motivated student to (1) learn about the MRI and signal processing theory behind this modality, (2) improve the current estimation methods, and (3) test and compare the new results with the current methods and histological measurements.
The project builds on top of previous cutting-edge research carried out in our lab (for more details see our multi-component T2 reconstruction toolbox and related references: https://github.com/ejcanalesr/multicomponent-T2-toolbox). The results will be published in international conferences and relevant journals.
Requirements: The project will be implemented in Python, so good knowledge is required. This project is ideal for a computer scientist, mathematician, physicist, or engineer interested in medical imaging, optimization, and signal processing.
Supervisors: Dr. Erick J. Canales-Rodríguez ([email protected]) and Prof. Jean-Philippe Thiran
5. Image analysis for cervical cancer detection
Cervical cancer is a major concern in public health around the world, both in high and low- and middle-income settings. In collaboration with the Geneva University Hospitals (HUG) and Dschang District Hospital in Cameroon, we aim at implementing a smartphone-based solution that automatically detects cervical cancer from videos of the cervix using deep neural networks.
Project 5.1 – Image segmentation and quality assessment for cervical cancer detection using deep learning
This project focuses on the preprocessing pipeline required before the classification of the images. Firstly, the goal is to improve the existing deep learning-based method for detecting and segmenting the cervix (region of interest) in the images. Then, based on those results, the second part addresses movement detection and compensation. Many further extensions of the project are possible in quality assessment of the images (blurriness, presence of blood, mucus or an object…).
Requirements: Basic knowledge of deep learning and image processing. Fluent in python and pytorch.
Project 5.2 Cervical cancer classification
Visual inspection with acetic acid is a common method used to detect cervical cancer. It consists of applying diluted acetic acid on the cervix which acts as a contrasting agent: the different types of tissues (and potential lesions) whiten at various rates and reach various intensity.
A deep learning model was trained to classify static images of the cervix based on images taken approximately 1 minute after application of the acetic acid. Firstly, this project aims to explore its performance on images taken at a different time after application of acetic acid in order to identify the optimal frame. A secondary objective is to improve the robustness of the model, e.g. training it with images at different times.
Requirements: Deep learning. Fluent in python and pytorch.
Assistants: Magali Jonnalagedda-Cattin ([email protected]) and Roser Vinals Terres ([email protected])
Supervisor: Prof. Jean-Philippe Thiran
6. Zero-/few-shot anomaly localization using a vision-language foundation model
Description: Anomaly detection and localization are the centerpieces of many safety-critical applications. For manufacturing defect detection, building a single model that can be generalized to various product types without or with only a few anomaly-free reference images is a promising research direction. This project aims to leverage recent open-source vision-language foundation models, such as the CLIP model [1], to perform anomaly segmentation in a zero-/few-shot setting. The developed method will be evaluated on various tasks related to the visual inspection of industrial images.
[1] Jeong, J., Zou, Y., Kim, T., Zhang, D., Ravichandran, A. and Dabeer, O. Winclip: Zero-/few-shot anomaly classification and segmentation. CVPR 2023
Requirements: Basic knowledge of deep learning. Fluent in Python and PyTorch.

Assistant: Dr Behzad Bozorgtabar ([email protected])
Supervisor: Prof. Jean-Philippe Thiran
In this project, we aim to gain a greater level of understanding of VLM’s learning process. To this end, we will investigate what features emerge during the training of VLM: are they specific to each modality – text or image – or do they capture the underlying common concepts depicted by both modalities? Do they show preferences for some features (e.g. removing “hard” features, capturing only “easy” common ones, a.k.a simplicity bias [5])?
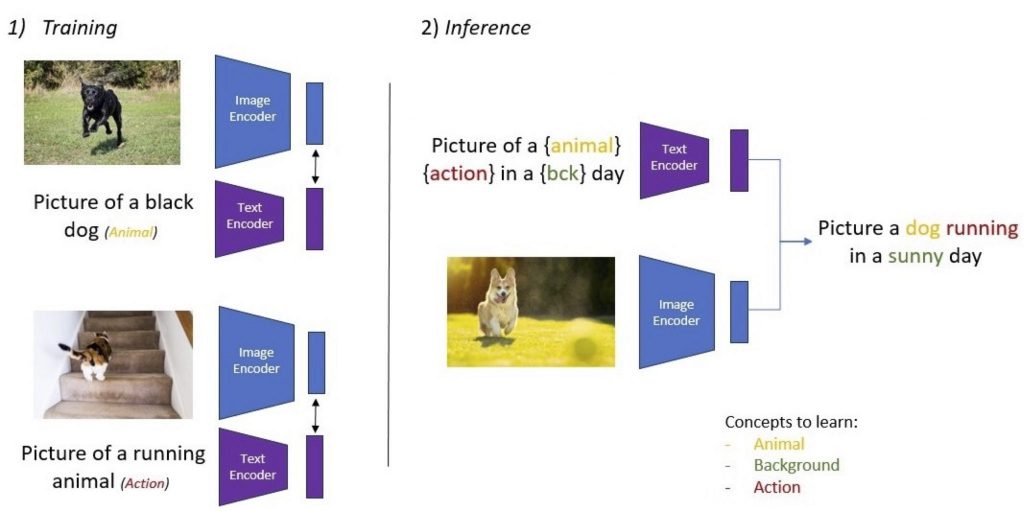
Fig. 1: What features are learned by vision-language models? When only action and animal concepts are described by the text, can we learn other visual concepts (e.g. background)? Can we compose the learned concepts (action + animal)?
To address these questions, a new synthetic VL dataset will be created, and several pre-trained models [1, 2, 3, 4] will be implemented and compared. The outcomes of these experiments could bring essential insights into what and how vision-language systems learn and how we could improve their learning process.
This project is an excellent opportunity to work at the edge between Computer Vision and Natural Language Processing problems and to gain a deeper understanding of current SOTA multi-modal models. This work will be done between ECEO and LTS5 laboratory.
Requirements:
– Strong Python programming skills
– Experience with Deep Learning libraries (TensorFlow or PyTorch)
– Familiarity with Computer Vision and/or Natural Language Processing
– Curiosity and eagerness to propose new solutions
Supervisors:
Dr. Benoit Dufumier ([email protected]), Prof. Jean-Philippe Thiran (EPFL Lausanne-LTS5)
Dr. Javiera Castillo Navarro (EPFL Valais-ECEO)
References
[1] ViLBERT: Pretraining Task-Agnostic Visiolinguistic Representations for Vision-and-Language Tasks, Lu et al., NeurIPS 2019
[2] Learning Transferable Visual Models From Natural Language Supervision, Radford et al., ICML 2021
[3] Vision-Language Pre-Training with Triple Contrastive Learning, Yang et al., CVPR 2022
[4] Learning Visual Representations via Language-Guided Sampling, El Banani et al., CVPR 2023
[5] The Pitfalls of Simplicity Bias in Neural Networks, Shah et al., NeurIPS 2020
MEDICAL IMAGING PROJECTS
1. Domain adaptation for ultrasound image quality enhancement with deep learning
This project focuses on improving the image quality of ultrasound images using a deep-learning-based image reconstruction techniques. The goal is to adapt the current CNN to a different setting and equipment configuration, enabling imaging at greater depths and wider fields of view. Various approaches, including transfer learning and training from scratch, will be explored using a new dataset. The project will be applied to fetal and liver imaging and 3D tracked systems.

To acquire the dataset we will ask the student to use an ultrasound device after proper training, potentially receiving compensation for creating a large in vivo dataset.
Requirements: Signal processing and deep learning. Fluent in python and pytorch.
Assistants: Roser Viñals Terres ([email protected]) and Sandra Marcadent ([email protected])
Supervisor: Prof. Jean-Philippe Thiran
2. Deep learning for foetal head segmentation on 3D-tracked ultrasound images
Due to its non-invasive nature, ultrasound imaging is a widely employed technique in the field of obstetrics and gynaecology. It relies on emitting ultrasound waves into tissues and receiving echoes produced at tissue interfaces.
By integrating ultrasonography with an optical tracking system, such as infrared cameras and a reflective marker on the ultrasound probe, 3D-tracked ultrasound imaging enables the continuous recording of the image plane’s position in 3D space. By employing this technique, we can reconstruct large volumes and achieve 4D visualization by fitting 3D models to the generated images and point clouds. In the frame of a larger project in obstetrics*, we aim to develop such visualization system for the foetal head during childbirth.
One crucial step towards this goal is the automatic and real-time segmentation of the foetal head on ultrasound images. The segmentation process would consist of two main parts. First, we need to extract a mask and biometric parameters from independent 2D images. Second, we need to accurately segment the fine contours of the skull from a set of 3D-tracked images to extract a 3D point cloud.

We are looking for a Master student to work on those two segmentation steps. The first part will involve the development and application of a deep learning approach, incorporating techniques such as transfer learning, data augmentation and the design of a task-specific training loss. For the second part, the student will develop a method utilizing the 3D spatial consistency amongst ultrasound images.
The ideal candidate for this project should have solid programming skills with proficiency in PyTorch and a strong foundation in image processing, machine learning and deep learning.
Assistants : Sandra Marcadent ([email protected]), Ayoub Tajja ([email protected])
Supervisor : Prof. Jean-Philippe Thiran
*You will find more information about it here: https://vimeo.com/321254812
3. Spherical Deconvolution Algorithms for Intra-Voxel Fiber Estimation and Brain Connectivity Mapping
The LTS5 Diffusion group focuses on brain tissue microstructure and structural connectivity –estimated by diffusion Magnetic Resonance Imaging (dMRI) data, with a particular focus on the reconstruction of the nerve fiber orientation distribution function (ODF) per voxel (see the figure below). This information is important for the reconstruction of the brain’s white matter by using fiber tracking algorithms (see ref [1]).

We have implemented various novel reconstruction algorithms (e.g., see refs. [2-5]) and we plan to develop a new generation of methods using Machine Learning techniques. The goals of this project are: (1) create a large database of fiber ODFs and corresponding dMRI signals, (2) Design, train, and optimize a neural network using this dataset, (3) predict the fiber ODFs from new dMRI data, and (4) compare the implemented algorithm with state-of-the-art techniques using both synthetic and real dMRI data acquired from human brains. The results will be published in international conferences and relevant journals.
Requirements: The project will be implemented in Python, so good knowledge is required. This project is ideal for a computer scientist, mathematician, physicist, or engineer interested in medical imaging, machine learning, signal processing, and optimization.
References:
[1] https://www.sciencedirect.com/science/article/abs/pii/S1053811914003541
[2] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4607500
[3] https://www.sciencedirect.com/science/article/abs/pii/S1053811918307699
[4] https://onlinelibrary.wiley.com/doi/10.1002/mrm.21917
Supervisors: Dr. Erick J. Canales-Rodríguez ([email protected]), Dr. Gabriel Girard ([email protected]), and Prof. Jean-Philippe Thiran
4. Myelin Water Imaging Using T2 Relaxometry
Myelin is a lipid-rich substance that surrounds the axons in the brain, which is essential for the proper functioning of the nervous system. Myelin water imaging is a magnetic resonance imaging (MRI) method that can be used to quantify and visualize myelination in the brain and spinal cord in vivo. The signal coming from the MRI machine (using a multi-echo T2 relaxometry sequence) can be decomposed into components, including that originated by water molecules trapped between the lipid bilayers of myelin. The correct estimation of this component provides a myelin-specific MRI biomarker to monitor brain changes in cerebral white matter. Myelin quantification has important implications for understanding various neurodegenerative diseases, including multiple sclerosis.
We are looking for a motivated student to (1) learn about the MRI and signal processing theory behind this modality, (2) improve the current estimation methods, and (3) test and compare the new results with the current methods and histological measurements.
The project builds on top of previous cutting-edge research carried out in our lab (for more details see our multi-component T2 reconstruction toolbox and related references: https://github.com/ejcanalesr/multicomponent-T2-toolbox). The results will be published in international conferences and relevant journals.

Requirements: The project will be implemented in Python, so good knowledge is required. This project is ideal for a computer scientist, mathematician, physicist, or engineer interested in medical imaging, optimization, and signal processing.
Supervisors: Dr. Erick J. Canales-Rodríguez ([email protected]) and Prof. Jean-Philippe Thiran
5. Image analysis for cervical cancer detection
Cervical cancer is a major concern in public health around the world, both in high and low- and middle-income settings. In collaboration with the Geneva University Hospitals (HUG) and Dschang District Hospital in Cameroon, we aim at implementing a smartphone-based solution that automatically detects cervical cancer from videos of the cervix using deep neural networks.
Project 5.1 – Image segmentation and quality assessment for cervical cancer detection using deep learning
This project focuses on the preprocessing pipeline required before the classification of the images. Firstly, the goal is to improve the existing deep learning-based method for detecting and segmenting the cervix (region of interest) in the images. Then, based on those results, the second part addresses movement detection and compensation. Many further extensions of the project are possible in quality assessment of the images (blurriness, presence of blood, mucus or an object…).
Requirements: Basic knowledge of deep learning and image processing. Fluent in python and pytorch.
Project 5.2 Cervical cancer classification
Visual inspection with acetic acid is a common method used to detect cervical cancer. It consists of applying diluted acetic acid on the cervix which acts as a contrasting agent: the different types of tissues (and potential lesions) whiten at various rates and reach various intensity.
A deep learning model was trained to classify static images of the cervix based on images taken approximately 1 minute after application of the acetic acid. Firstly, this project aims to explore its performance on images taken at a different time after application of acetic acid in order to identify the optimal frame. A secondary objective is to improve the robustness of the model, e.g. training it with images at different times.
Requirements: Deep learning. Fluent in python and pytorch.
Assistants: Magali Jonnalagedda-Cattin ([email protected]) and Roser Vinals Terres ([email protected])
Supervisor: Prof. Jean-Philippe Thiran
6. Deep learning based shape analysis of cardiac biventricular meshes – collaboration with CHUV
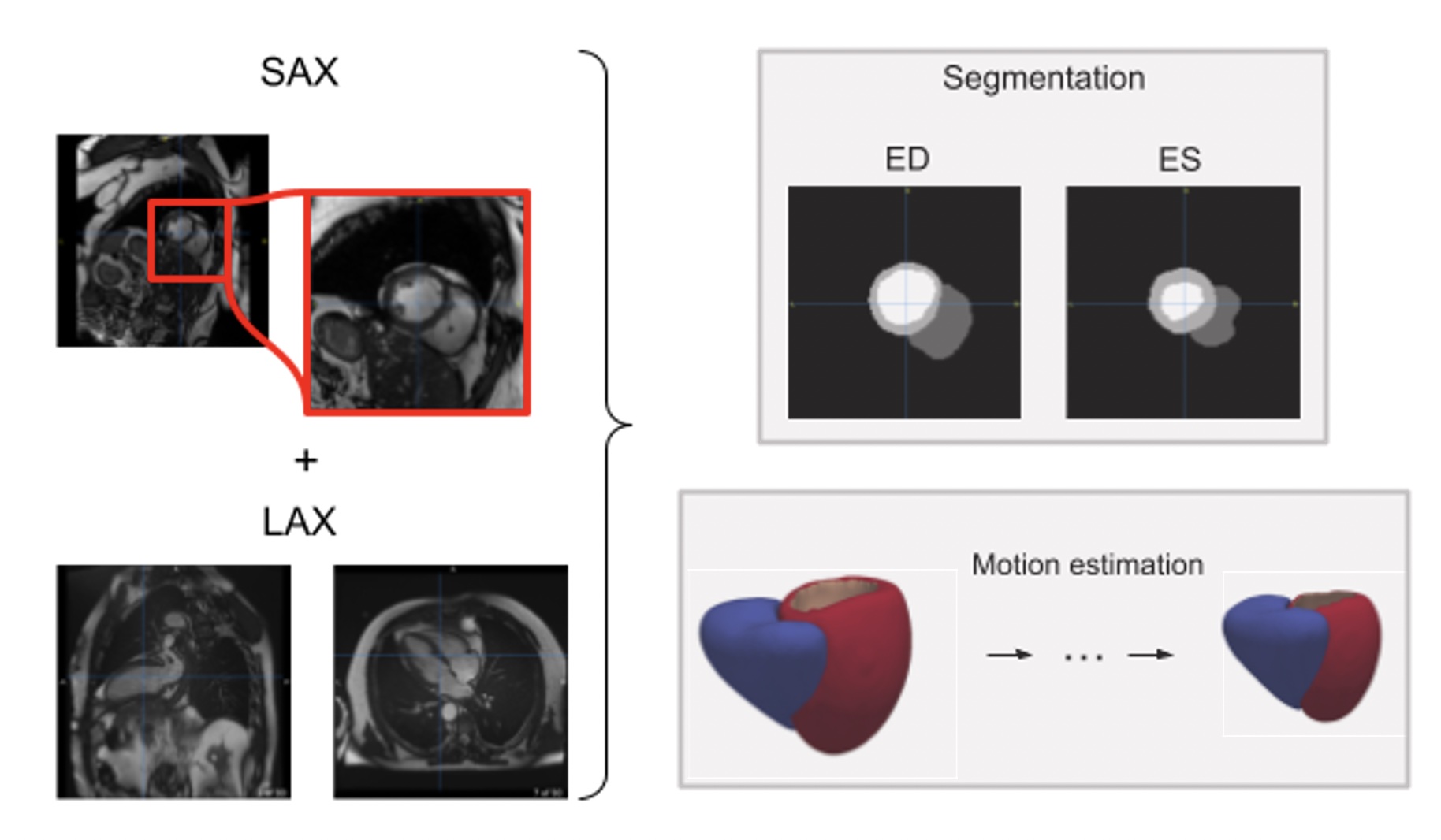
The goal of the project is to make the student familiar with the current trends in medical cardiac imaging analysis. Cardiac motion and shape analysis have proved to be useful to characterize differences in clinical diseases [1,2]. However, the pipeline to obtain suitable meshes from cardiac magnetic resonance images is relatively complex and the displacement estimation relies on single image modalities with a single point of view. Recent work focused on the integration of multiple cardiac image modalities covering different points of view to jointly predict mesh displacements throughout the cardiac cycle [3]. Nevertheless, the exploration of how useful mesh descriptors can be in large-scale datasets remains relatively unexplored [2]. In this project, the student will use state-of-the-art deep learning approaches based on differential geometry and graph neural networks to explore the potential of shape descriptors to stratify subjects by clinical diagnosis in a large-scale cohort, the UK Biobank, containing thousands of cardiac images.
Therefore, the goals are 1) apply existing meshing methods to reliably obtain biventricular meshes [4,5] from multi-structure segmentations obtained from previous works based on convolutional neural networks (CNN) [6,7], 2) generate a statistical shape atlas to study the main modes of variation [8] and 3) explore the use of state-of-the art shape analysis tools to characterize the cardiac shape of each subject [9]
The project will provide valuable input to an ongoing research effort between Lausanne and Geneva in integrative characterisation of heart failure, and therefore has the potential to contribute to advancing medical science and ultimately benefit patients with cardiac pathologies.
References:
[1]: Mansi, T., et al..: A statistical model for quantification and prediction of cardiac remodelling: Application to Tetralogy of Fallot. IEEE Trans Med Imaging
[2]: Bello, G. A.,et al. (2019). Deep-learning cardiac motion analysis for human survival prediction. Nature Machine Intelligence
[3]: Meng, Q., Bai, W., Liu, T., O’Regan, D. P., & Rueckert, D. (2022). Mesh-Based 3D Motion Tracking in Cardiac MRI Using Deep Learning, MICCAI 2022
[4]: Wickramasinghe, U., Remelli, E., Knott, G., & Fua, P. (2020). Voxel2Mesh: 3D Mesh Model Generation from Volumetric Data. MICCAI 2020
[5]: William E. Lorensen and Harvey E. Cline. 1987. Marching cubes: A high resolution 3D surface construction algorithm. SIGGRAPH Comput. Graph. 21, 4 (July 1987)
[6]: Bai, W., et al. (2018). Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. Information and Computing Sciences. Artificial Intelligence and Image Processing. Journal of Cardiovascular Magnetic Resonance, 20(1).
[7]: Byrne, N., Clough, J. R., Valverde, I., Montana, G., & King, A. P. (2022). A persistent homology-based topological loss for CNN-based multi-class segmentation of CMR. IEEE Transactions on Medical Imaging.
[8]: Bai, W., et al. (2015). A bi-ventricular cardiac atlas built from 1000+ high resolution MR images of healthy subjects and an analysis of shape and motion. Medical Image Analysis
[9]:Sharp, N., Attaiki, S., Crane, K., & Ovsjanikov, M. (2022). DiffusionNet: Discretization Agnostic Learning on Surfaces. ACM Transactions on Graphics, 41(3), 1–16.
Requirements: The project will be implemented in Python. Good knowledge of Pytorch as well as familiarity with deep learning are desirable.
Supervisor:
Prof. Jean-Philippe Thiran (EPFL-LTS5)
Co-supervisors:
Dr. Jaume Banus Cobo CHUV-Translational Machine Learning Lab ([email protected])
Dr. Jonas Richiardi CHUV-Translational Machine Learning Lab ([email protected])
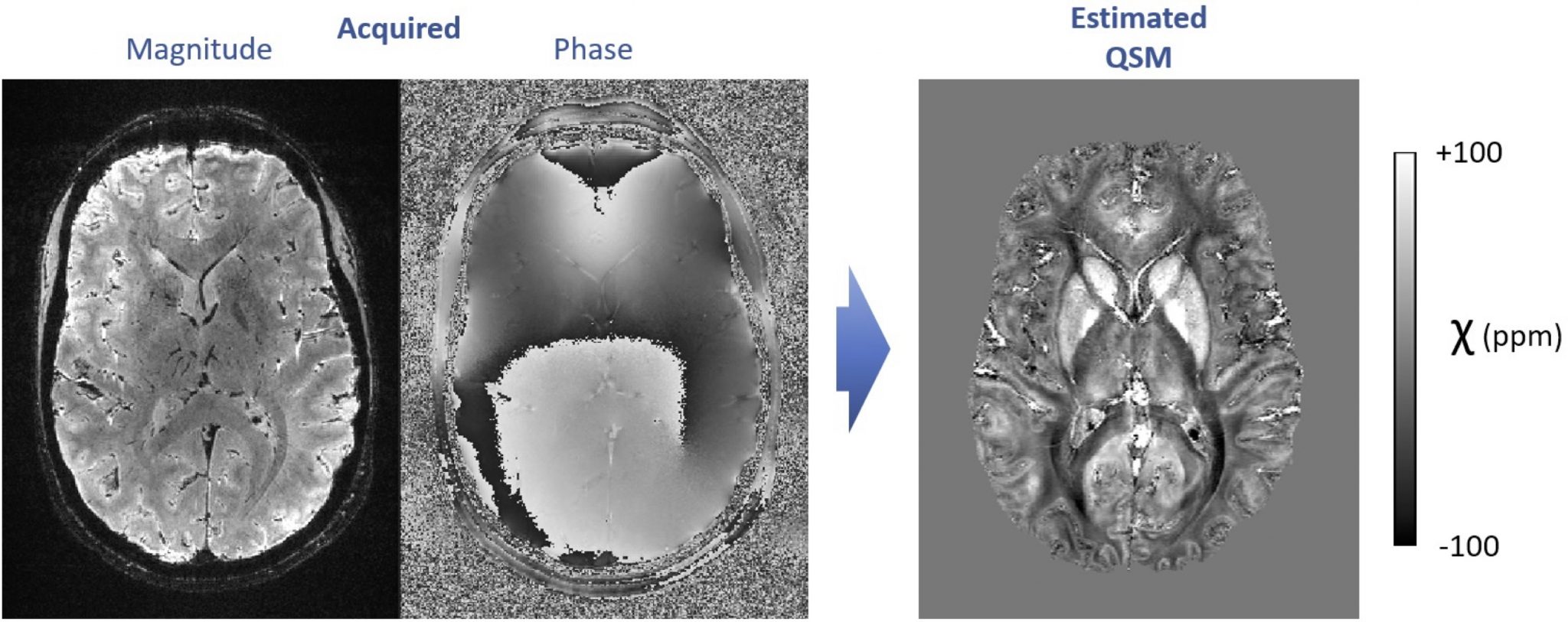
7. High-resolution quantitative susceptibility mapping with 7T MRI
Using dedicated image acquisition and processing techniques, MRI allows mapping the distribution of magnetic susceptibility in living tissues, non-invasively and without injected contrasts. Magnetic susceptibility is currently under research for numerous clinical applications such as the detection of microbleeds, calcifications and abnormal iron metabolism, in the brain as well as other organs.
We work on improving quantitative susceptibility mapping (QSM) approaches at 7 Tesla, to generate exquisitely detailed, sub-millimeter resolution images. To achieve this goal, the topics of interest include the mitigation of interference from motion, the reduction of effects from blood vessels and blood flow, and the improvement of QSM reconstruction techniques based on acquired field maps (ill-posed inverse problem). The solutions include cutting-edge approaches in model-based image processing and/or deep learning.
Within this range of topics, diverse MSc projects can be planned and flexibly adapted to the student’s background and interests. Motivated students are encouraged to contact us to discuss available projects. These projects are held in collaboration with CSEM.
Ideal requirements:
- Interest in brain imaging
- Good knowledge in Python
- Knowledge of image processing fundamentals
- Fluent in written and spoken English
Supervisors:
Dr. João Jorge – [email protected] – CSEM
Dr. Meritxell Bach Cuadra – [email protected] – CIBM SP CHUV-UNIL
Prof. Jean-Philippe Thiran – [email protected]
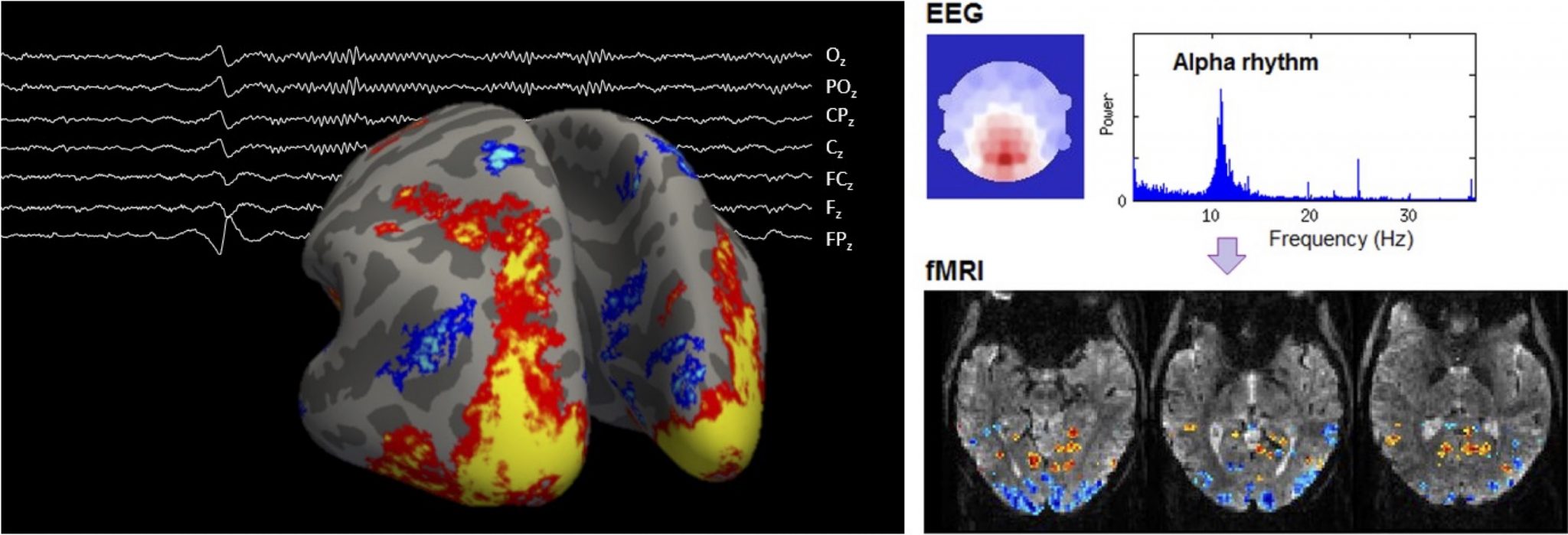
8. Multimodal imaging of brain function at ultra-high spatial & temporal resolution with combined EEG-fMRI at 7T
Electroencephalography (EEG) and functional magnetic resonance imaging (fMRI) are valuable brain imaging tools that can detect, respectively, electrical and vascular changes that occur during brain function, non-invasively. We work on the combination of these techniques at a magnetic field of 7T, where fMRI has strong boosts in sensitivity, enabling unprecedented levels of spatial and temporal specificity. Within this domain, several lines of research are open for study :
– Methodology-oriented: Both EEG and fMRI can be affected by important degradation effects when acquired together, especially at 7T. Dedicated improvements at the level of the signal acquisition and/or signal processing and analysis are vital to obtain high-quality data.
– Application-oriented: Our unique, optimized datasets allow studying subtle features of brain function such as thalamocortical interactions and cortical layer-specific activity, which remain largely unexplored. This can be investigated using cutting-edge signal processing and analysis techniques, to be adapted to these unique datasets.
Within this range of topics, MSc projects can be planned and flexibly adapted to the student’s background and interests. Motivated students are encouraged to contact us to discuss available projects. These projects are held in collaboration with CSEM.
Ideal requirements:
- Interest in brain imaging
- Good knowledge in Python
- Knowledge of signal and/or image processing fundamentals
- Fluent in written and spoken English
Supervisors:
Dr. João Jorge – [email protected] – CSEM
Dr. Meritxell Bach Cuadra – [email protected] – CIBM SP CHUV-UNIL
Prof. Jean-Philippe Thiran – [email protected]
9. Explainable AI for the Detection of Rim Lesions in 3D MRI Scans
This project aims to contribute to the development of artificial intelligence-aided (AI-aided) tools for the prognosis of multiple sclerosis (MS) disease in clinical practice. The focus of this project is on the detection of rim lesions in MS, which are specific patterns of brain lesions visible on magnetic resonance imaging (MRI) scans [1]. Rim lesions are believed to indicate ongoing inflammation and active demyelination, particularly in the outer edges of pre-existing lesions.
The current manual process of detecting rim lesions in MRI scans is time-consuming and requires significant expertise. Therefore, automating this task through deep learning (DL) techniques, specifically 3D semantic segmentation, appears to be a promising solution [2]. However, due to the inherent noise in ground truth segmentation masks for both lesion detection and delineation, employing weakly supervised segmentation (WSS) methods becomes an attractive alternative. In this project, we will explore the use of saliency detection methods for weakly supervised segmentation purposes [3]. Thus, the student project will encompass two main stages: 1. Creating a DL model for a preliminary regression task of the number of lesions in a scan. 2. Applying Explainable AI (XAI) techniques, particularly saliency maps [4], for the detection of rim lesions.
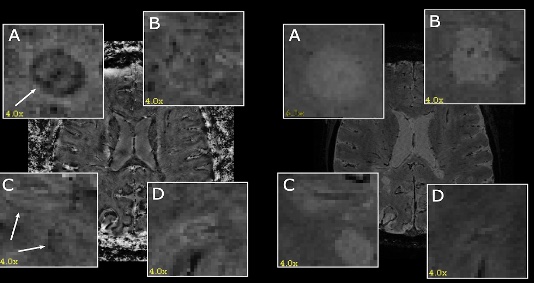
Requirements for this project include experience in deep learning with Python (PyTorch) and experience in image/signal processing.
The project will be supervised by Prof. Jean-Philippe Thiran (EPFL), with co-supervision from Nataliia Molchanova ([email protected]) (CHUV, HESSO) and Meritxell Bach Cuadra ([email protected]) (CIBM).
References:
[1] https://onlinelibrary.wiley.com/doi/pdf/10.1002/ana.25877? casa_token=j0AshM4gBNgAAAAA:3pYQW2ovUNtxSBoWZLaQklEHmsU1muopaB4WirKr5zYaTsf NioNkySneboE-VJF91mUkcjvnWtFMDgUnjg
[2] https://www.sciencedirect.com/science/article/pii/S2213158220301728
[3] https://openaccess.thecvf.com/content_ICCV_2019/papers/ Zeng_Joint_Learning_of_Saliency_Detection_and_Weakly_Supervised_Semantic_Segmentation_I CCV_2019_paper.pdf
[4] https://pubs.rsna.org/doi/pdf/10.1148/ryai.2021200267
COMPUTER VISION PROJECTS
10. Zero-/few-shot anomaly localization using a vision-language foundation model
Description: Anomaly detection and localization are the centerpieces of many safety-critical applications. For manufacturing defect detection, building a single model that can be generalized to various product types without or with only a few anomaly-free reference images is a promising research direction. This project aims to leverage recent open-source vision-language foundation models, such as the CLIP model [1], to perform anomaly segmentation in a zero-/few-shot setting. The developed method will be evaluated on various tasks related to the visual inspection of industrial images.
[1] Jeong, J., Zou, Y., Kim, T., Zhang, D., Ravichandran, A. and Dabeer, O. Winclip: Zero-/few-shot anomaly classification and segmentation. CVPR 2023
Requirements: Basic knowledge of deep learning. Fluent in Python and PyTorch.

Assistant: Dr Behzad Bozorgtabar ([email protected])
Supervisor: Prof. Jean-Philippe Thiran

Fig. 1: What features are learned by vision-language models? When only action and animal concepts are described by the text, can we learn other visual concepts (e.g. background)? Can we compose the learned concepts (action + animal)?
To address these questions, a new synthetic VL dataset will be created, and several pre-trained models [1, 2, 3, 4] will be implemented and compared. The outcomes of these experiments could bring essential insights into what and how vision-language systems learn and how we could improve their learning process.
This project is an excellent opportunity to work at the edge between Computer Vision and Natural Language Processing problems and to gain a deeper understanding of current SOTA multi-modal models. This work will be done between ECEO and LTS5 laboratory.
Requirements:
– Strong Python programming skills
– Experience with Deep Learning libraries (TensorFlow or PyTorch)
– Familiarity with Computer Vision and/or Natural Language Processing
– Curiosity and eagerness to propose new solutions
Supervisors:
Dr. Benoit Dufumier ([email protected]), Prof. Jean-Philippe Thiran (EPFL Lausanne-LTS5)
Dr. Javiera Castillo Navarro (EPFL Valais-ECEO)
References
[1] ViLBERT: Pretraining Task-Agnostic Visiolinguistic Representations for Vision-and-Language Tasks, Lu et al., NeurIPS 2019
[2] Learning Transferable Visual Models From Natural Language Supervision, Radford et al., ICML 2021
[3] Vision-Language Pre-Training with Triple Contrastive Learning, Yang et al., CVPR 2022
[4] Learning Visual Representations via Language-Guided Sampling, El Banani et al., CVPR 2023
[5] The Pitfalls of Simplicity Bias in Neural Networks, Shah et al., NeurIPS 2020
12. Fall detection using machine learning – master project in industry at Gets MSS SA (Lausanne)
At Gets MSS Sa, we aim to make the work of caregivers easier while improving security of patients and elderly people. To achieve that we develop and manufacture nursecall system that allow caregiver to be alerted when a patient needs help.
Falls are a problem encountered when someone is too weak to be able to get up again or to move around alone. Therefore to be able to give the alert in case a person has fallen, we would like to develop a fall detection system. We aim to be able to know when a person goes out of bed to prevent falls.
We need you for:
- Review existing technologies to be able to detect those falls. For example technologies radar, Lidar, Depth of Field, Kinect, etc…
- Identify the advantages and disadvantages of each technology.
- Develop a prototype using the chosen technology, image processing and machine learning.
We would like to create this product through an embedded system that will respect privacy.
Gets MSS SA is a small company of about 30 employees with a family atmosphere. Strong of its development team, it has become the leader in the Swiss nursecall market, maintaining itself on the cutting edge of innovation.
Contact : [email protected] and Prof. J.-Ph. Thiran