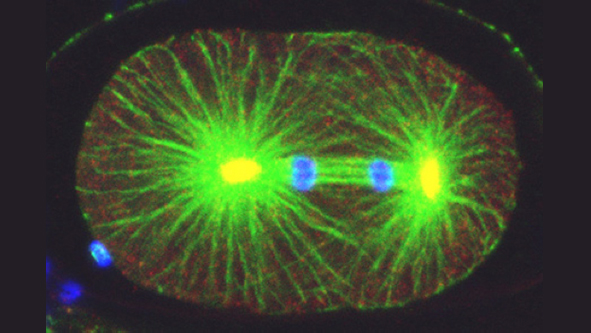
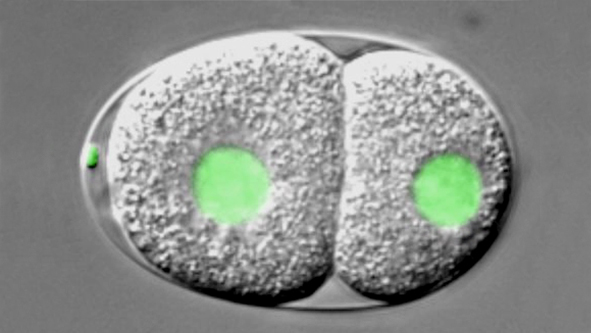
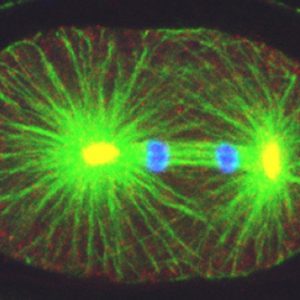
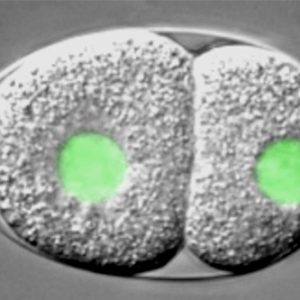
How living systems break symmetry in an organized manner is an important question in biology. One instance where this takes place is during asymmetric division, a process crucial for generating cellular diversity in development and in stem cell lineages. In this framework, we uncovered recently that the C. elegans zygote possesses self-organizing properties that result in autonomous polarization. Moreover, we found that the kinase Aurora A normally plays a critical role in funneling those properties onto a single site, next to paternally contributed centrioles, thus ensuring spatial coupling between paternal and maternal contributions at development onset. After polarity establishment along the anterior-posterior (A-P) embryonic axis of the C. elegans zygote, the mitotic spindle becomes positioned eccentrically, thus directing cleavage into two cells that differ in size and fate. Our previous work indicates that such spindle positioning requires the function of an evolutionary conserved ternary complex that anchors the minus end directed motor protein complex dynein at the cell cortex. There, dynein is thought to generate pulling forces on astral microtubules that emanate from the spindle poles, thus positioning the mitotic spindle.
Watch movie of C. elegans embryo carrying a GFP-histone2B fusion protein imaged using dual time-lapse differential interference contrast (DIC) and overlaid fluorescence microscopy, from the one-cell stage until the four-cell stage. The embryo is ~50μm long and the movie is shown at ~100 times speed of the actual processes.
Watch movie of C. elegans one-cell embryo expressing RFP::NMY-2 (red); GFP::PAR-2 and GFP::SAS-7 (both green). Differential interference contrast (DIC) image is overlaid onto the fluorescence channels. The embryo is ~50 μm long and time is shown in min:sec. From Klinkert et al., 2019.
Watch movie of C. elegans one-cell air-1(RNAi) embryo expressing RFP::NMY-2 (red); GFP::PAR-2 and GFP::SAS-7 (both green). Differential interference contrast (DIC) image is overlaid onto the fluorescence channels. The embryo is ~50 μm long and time is shown in min:sec. From Klinkert et al., 2019.