Laboratory of Therapeutic Proteins and Peptides
Open Positions:
Open PhD position: We have a new PhD position for a project in which we want to develop membrane-permeable cyclic peptides for addressing intracellular proteins. This position is one of 17 positions of the MSCA Doctoral Network “MC4DD – Macrocycles for Drug Discovery“
Note: for this PhD position in the Heinis lab, the funding instrument requires that candidates must not have resided or carried out their main activity (work, studies, etc.) in Switzerland for more than 12 months in the 3 years immediately before the recruitment date. Find more information here. For interested students, please send a CV and cover letter to Prof. Christian Heinis. Please find more PhD positions funded by the same
PhD projects: We expect to soon open new PhD positions in cyclic peptide drug development (combinatorial chemistry, HTS, drug development). For interested students, please send a CV and cover letter to Prof. Christian Heinis.
Master projects: We are looking for enthusiastic master students. Candidates are invited to send their CV via email to Prof. Christian Heinis.
Currently we focus on the following three projects:
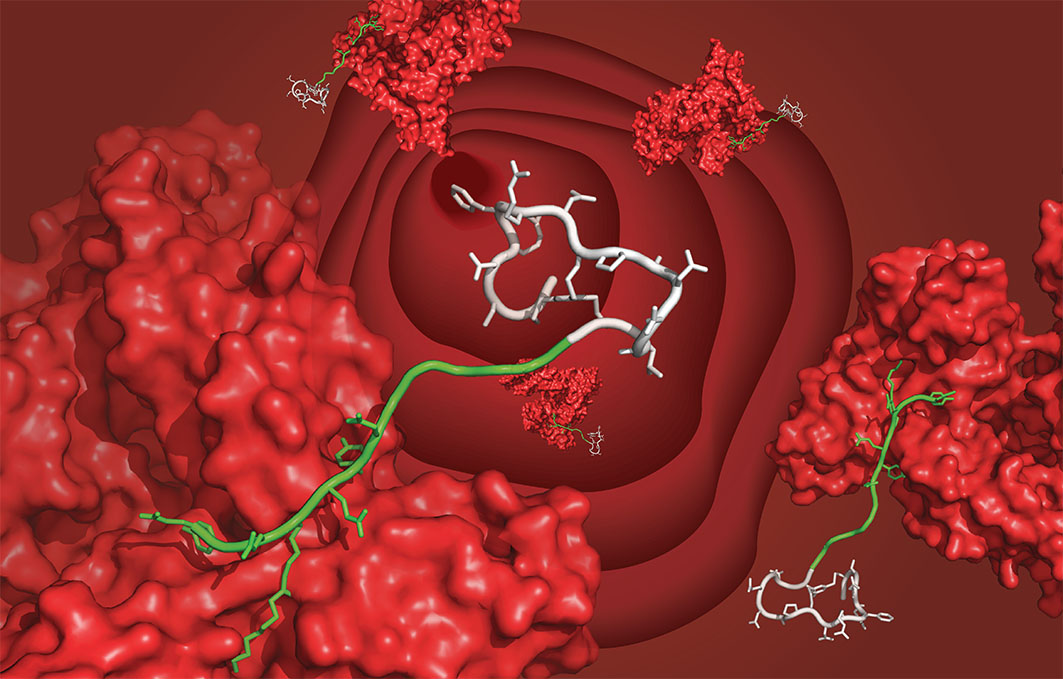
Development of drugs based on cyclic peptides
The ultimate goal of our laboratory is the development of therapeutics for addressing unmet medical needs. We work with cyclic peptides because they can engage with difficult protein targets to which classical small molecules can hardly bind.
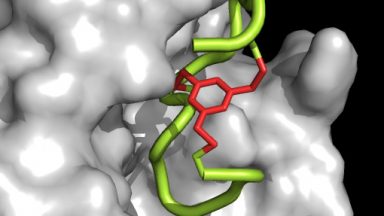
Phage display selection of bicyclic peptides
Phage display technology allows for the genetic encoding of billions of different peptide sequences. We combine phage display and chemical reactions to generate and screen large combinatorial libraries of bicyclic peptides.

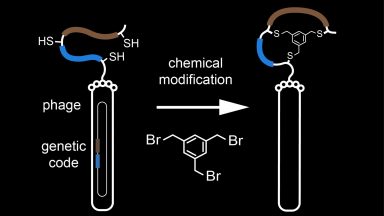
High-throughput synthesis of macrocycles
A major goal of our laboratory is the development of cell permeable or even orally available macrocycles that bind to targets of interest. Towards this end, we develop methods for the combinatorial synthesis of large libraries of sub-kDa macrocycles.
Selected recent publications
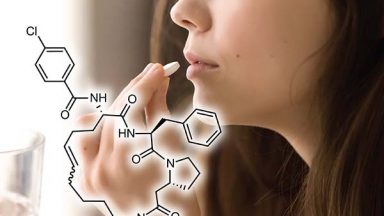
De novo development of small cyclic peptides that are orally bioavailable
Merz, M.L., Habeshian, S., Li, B., David, J.-A.G.L., Nielsen, A.L., Ji, X., Il Khwildy, K., Duany Benitez, M.M., Phothirath, P. and Heinis, C.
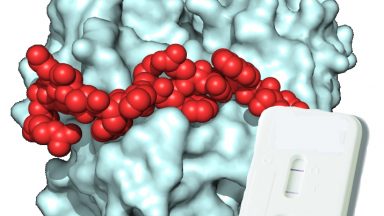
High-affinity peptides developed against calprotectin and their application as synthetic ligands in diagnostic assays
Díaz-Perlas, C., Ricken, B., Farrera-Soler, L., Guschin, D., Pojer, F., Lau, K., Gerhold, C.-B. and Heinis, C.

Synthesis and direct assay of large macrocycle diversities by combinatorial late-stage modification at picomole scale
Habeshian, S., Merz, M.L., Sangouard, G., Mothukuri, G.K., Schüttel, M., Bognár; Z., Díaz-Perlas, C., Vesin, J., Bortoli Chapalay, J., Turcatti, G., Cendron, L., Angelini, A. and Heinis, C.
We are grateful for support by the Swiss National Science Foundation and the European Research Council.

