Besides the below-offered topics for master and semester projects, we welcome you to bring your ideas and elaborate on a project with us.
Whether you are interested in one listed topic or bring some ideas, we strongly encourage you to meet with lab members to learn about the research in LCSB.
To get the most out of your internship or master’s project, we recommend reading the following article:
Ten simple rules for getting the most out of a summer laboratory internship
Interpretability studies of Deep Learning Methods for understanding the dynamic properties of metabolic networks
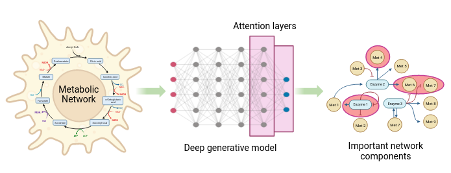
Generating large omics datasets has become routine practice to gain insights into cellular processes, yet deciphering such massive datasets and determining intracellular metabolic states remains challenging. Kinetic models of metabolism play a critical role in integrating omics data, as they provide explicit connections between metabolite concentrations, metabolic fluxes, and enzyme levels. However, the difficulties in determining kinetic parameters that govern cellular physiology hinder the broader adoption of these models by the research community. In this project, we aim to gain deeper insights into the relationship between the kinetic parameters of a metabolic network and the resulting dynamic properties. The student will perform interpretability studies on neural networks that have been already trained to generate kinetic models from scratch using recently developed Machine learning-based methods (RENAISSANCE1, REKINDLE2) or classify/predict the dynamic properties of large-scale metabolic models.
i) Generation: For generative deep learning methods, implementing attention layers during optimization can be useful for looking at important features. Attention mechanisms3 reveal the features or combination of features that are essential for completing the deep-learning objective and, hence, can reveal the intrinsic network-level correlations between kinetic parameters of metabolic networks and emergent dynamic properties.
ii) Classification/Prediction: Once a neural network has already been trained (>90% accuracy) to predict/classify the dynamic properties of a metabolic model, one can use recently established interpretability (grey box) techniques like Integrated Gradients, Neural Additive Models, Concept Activation Vectors, etc to get an idea of the influential features of the network that dictates the dynamics. All of these methods can be easily installed and implemented in Python through libraries like Captum (https://github.com/pytorch/captum), and Foolbox (https://github.com/bethgelab/foolbox).
| Type: | Master or semester project |
| Keywords: | Machine learning, neural networks, kinetic models, metabolism |
| Recommended knowledge: | Basics of Chemistry/Chemical engineering/Biology, Data analysis |
| Suggested reading: |
Choudhury, S., Narayanan, B., Moret, M., Hatzimanikatis, V., & Miskovic, L. (2023). Generative machine learning produces kinetic models that accurately characterize intracellular metabolic states. bioRxiv, 2023-02. Choudhury, S., Moret, M., Salvy, P., Weilandt, D., Hatzimanikatis, V., & Miskovic, L. (2022). Reconstructing kinetic models for dynamical studies of metabolism using generative adversarial networks. Nature Machine Intelligence, 4(8), 710-719. Niu, Z., Zhong, G., & Yu, H. (2021). A review on the attention mechanism of deep learning. Neurocomputing, 452, 48-62. |
| Contact: | S. Choudhury, L. Miskovic |
Using deep evolutionary algorithms to uncover potential regulatory mechanisms in metabolic networks for non-model organisms
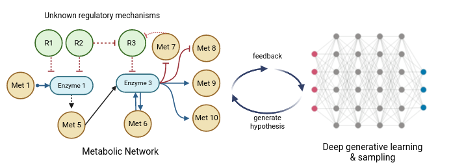
Metabolic regulation plays a crucial role in determining the overall efficiency and health of a cell1. Regulation provides essential biochemical feedback mechanisms that strongly affect physiological state of a cell. Metabolic enzyme kinetics usually involves complex interaction between many different molecules through allosteric interventions. Even though these interactions have been well studied for model organisms (like E. coli, S. cerevisiae, H. sapiens), these mechanisms and the regulatory candidates remain largely unknown for other less studied organisms. Recently, this has been attempted using Bayesian approaches and ensemble modelling2. In this project we propose a more generalized approach to uncover regulatory candidates by including all suspected candidates of regulation from literature. We then sample kinetic parameter sets (Km, Ki, wi) where Kms are the concentration associated Michaelis Menten constants, Kis are the regulatory constants and wis are the weights of regulation for the ith regulatory candidate, very efficiently using RENAISSANCE3. Then, the samples will be analysed to determine the feasibility of the proposed regulatory mechanism. This will involve setting up conditions for pruning the sampled parameter sets to remove biochemical, stoichiometric & thermodynamic infeasibilities. In light of the multiplicity of the solutions, this step will be crucial for the success of the project and hence needs more refinement. Also, the manner of sampling needs to be studied as it might be more computationally optimal to do the sampling in steps (first sampling the Kms and then the Kis and wi, or other alternatives).
| Type: | Master or semester project |
| Keywords: | Machine learning, neural networks, kinetic models, metabolism |
| Recommended knowledge: | Basics of Chemistry/Chemical engineering/Biology, Data analysis |
| Suggested reading: |
Metallo, Christian M., and Matthew G. Vander Heiden. “Understanding metabolic regulation and its influence on cell physiology.” Molecular cell 49.3 (2013): 388-398. Saa, Pedro A., and Lars K. Nielsen. “Construction of feasible and accurate kinetic models of metabolism: A Bayesian approach.” Scientific reports 6.1 (2016): 29635. Choudhury, S., Narayanan, B., Moret, M., Hatzimanikatis, V., & Miskovic, L. (2023). Generative machine learning produces kinetic models that accurately characterize intracellular metabolic states. bioRxiv, 2023-02. |
| Contact: | S. Choudhury, I. Toumpe, L. Miskovic |
Reconstruction and Exploration of the Bee Gut Microbial Community
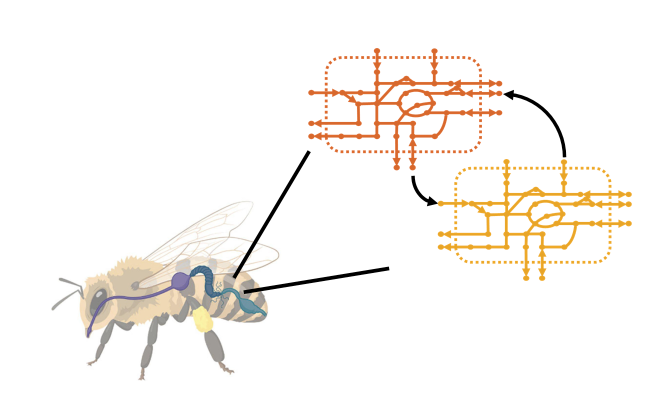 Bees are at the epicentre of our complex ecosystem. Consequently, the significant increase in fatalities of this vital pollinator raises great concerns about the reproduction of plants and the agricultural production on which humans depend. Numerous studies have shown that bacterial communities (called microbiota) composition and interactions in animals’ guts are critical for their hosts’ general health. Therefore, understanding the bees’ microbiota interactions would allow us to apprehend the issues behind bees decline.
Bees are at the epicentre of our complex ecosystem. Consequently, the significant increase in fatalities of this vital pollinator raises great concerns about the reproduction of plants and the agricultural production on which humans depend. Numerous studies have shown that bacterial communities (called microbiota) composition and interactions in animals’ guts are critical for their hosts’ general health. Therefore, understanding the bees’ microbiota interactions would allow us to apprehend the issues behind bees decline.
The reconstruction of high-quality curated genome-scale metabolic models (GEMs) of bee gut bacteria aims to help understand the complex interactions within the microbiota. GEM’s reconstruction allows the application of modern Systems Biology tools for analyzing and understanding behaviour under different environmental conditions.
Our group, Laboratory of Computational System Biotechnology (LCSB), is part of a Federal competence centre on microbiomes (https://nccr-microbiomes.ch/) working on experimental and computational approaches to model, analyze, and engineer natural and synthetic microbiota in animals, plants and environment. This master project will be part of one of these collaborations with Prof Philipp Engel at the University of Lausanne (https://engelbeelab.com/).
Within this project, we offer two master projects. One is the exploration of the metabolic capabilities of two organisms for which the genome-scale models have already been reconstructed. The second one is the reconstruction of the other organisms populating the bee gut microbiome. Both projects will start with the students familiarising themselves with the in-house methods and models. Then, the students will perform a literature search on relevant scientific achievements around the bee gut microbiota and identify the open questions about the microbiota’s complex interactions, such as the effects of pesticide or secondary bacteria, foreign bacteria, and variation of the nutrition or others. Finally, the students will use the available tools and the information collected to answer open questions and others that might arise during the project.
| Type: | Master or semester project |
| Keywords: | Genome-Scale Metabolic Models, Bee Gut Microbiota, Metabolism |
| Recommended knowledge: | Chemistry/Chemical engineering, Bio-Informatics, Matlab or Python |
| Suggested reading: | Raymann, K. & Moran, N. A. The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect Sci. 26, 97–104 (2018). Kwong, W. K., Engel, P., Koch, H. & Moran, N. A. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc. Natl. Acad. Sci. 111, 11509–11514 (2014). Vayena, E. et al. A workflow for annotating the knowledge gaps in metabolic reconstructions using known and hypothetical reactions. Proc. Natl. Acad. Sci. 119, e2211197119 (2022). |
| Contact: | E. Patelli, V. Hatzimanikatis |
Dynamic studies of metabolism using Machine Learning
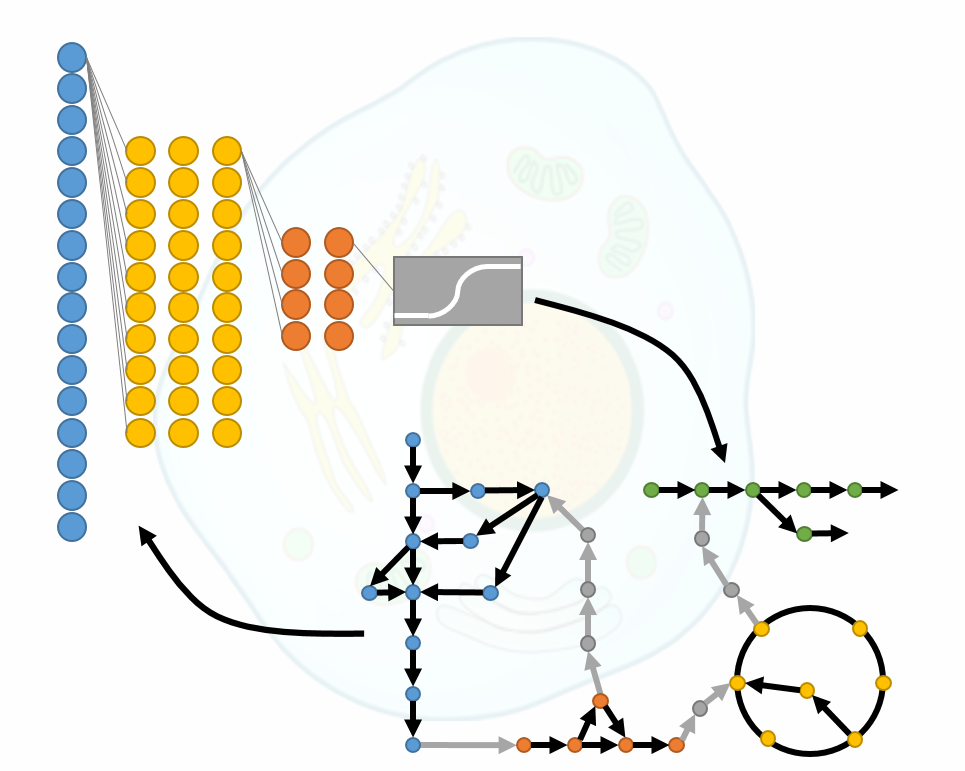 Kinetic models are indispensable in systems and synthetic biology studies of metabolism as they allow us to capture the dynamic behavior of metabolism and predict dynamic responses of living organisms to genetic and environmental changes. The scientific community has recognized the utility and potential of kinetic models, and efforts toward building large-scale and genome-scale kinetic models are recently intensified. Nevertheless, the development of these models is still facing the challenges, such as the characterization of the parameter space. Despite the abundantly available data, it is difficult to determine the parameters accurately describing the observed physiology. The reason for this is intrinsic nonlinearities and complex dynamic behavior of living organisms.
Kinetic models are indispensable in systems and synthetic biology studies of metabolism as they allow us to capture the dynamic behavior of metabolism and predict dynamic responses of living organisms to genetic and environmental changes. The scientific community has recognized the utility and potential of kinetic models, and efforts toward building large-scale and genome-scale kinetic models are recently intensified. Nevertheless, the development of these models is still facing the challenges, such as the characterization of the parameter space. Despite the abundantly available data, it is difficult to determine the parameters accurately describing the observed physiology. The reason for this is intrinsic nonlinearities and complex dynamic behavior of living organisms.
In this Master’s project, we propose to address this problem by studying generative neural networks to construct a population of large-/genome- scale kinetic models consistent with the experiments. The project will be conducted in several phases: training the neural network, validation of results, sensitivity analysis/hyperparameter tuning, and application to a broader class of problems. The obtained models will then be validated against the experimental data. Then, the student will improve the neural network’s performance by analyzing its structure. Finally, we will explore the application of such fine-tuned neural networks to a set of experimental data that describes alternative physiologies.
| Type: | Master or semester project |
| Keywords: | Machine learning, neural networks, kinetic models, metabolism |
| Recommended knowledge: | Basics of Chemistry/Chemical engineering/Biology, Data analysis |
| Suggested reading: | Choudhury et al., Reconstructing Kinetic Models fro Dynamical Studies of Metabolism Using Generative Adversarial Networks, Nature Machine Intelligence, (4), pp 710-719 (2022). Miskovic & Hatzimanikatis, Production of biofuels and biochemicals: in need of an ORACLE, Trends in Biotechnology, 8(28), pp 391-397 (2010). |
| Contact: | S. Choudhury, L. Miskovic |
Cell culture optimization and production of biopharmaceuticals using a CHO genome-scale model.
 Biopharmaceuticals are medical drugs produced through genetic manipulation of living cells. They are mainly produced using mammalian cell lines given their similitude with human cells. The Chinese hamster ovary (CHO) are the most used cell lines to produce therapeutic proteins.
Biopharmaceuticals are medical drugs produced through genetic manipulation of living cells. They are mainly produced using mammalian cell lines given their similitude with human cells. The Chinese hamster ovary (CHO) are the most used cell lines to produce therapeutic proteins.
The reconstruction of high-quality curated genome scale metabolic models (GEMs) for CHO cells, enables the application of modern Systems Biology for the analysis and understanding of culture behavior under different bioprocessing conditions. The consideration of constraint-based mathematical models facilitates the integration of available experimental data to analyze and understand the relationships between the different elements of the system and predict the response to perturbations.
The objective of this project is to study CHO cells metabolism under different environmental and bioprocessing conditions (media composition, feeding schedule). We will use the CHO GEM and constraint-based methods such as flux balance analysis or thermodynamic-base balance analysis to examine the alternative culture media, to study the effect of the medium composition on intracellular reactions, and to reveal metabolic differences among cell lines.
| Type: | Master or semester project |
| Keywords: | CHO genome scale, cell culture optimization, biopharmaceuticals |
| Recommended knowledge: | Chemistry/Chemical engineering, Bio-Informatics |
| Suggested reading: | Rejc Z, et al. Computational modelling of genome-scale metabolic networks and its application to CHO cell cultures. Computers in Biology and Medicine 88 (2017) 150–160 H. Hefzi, et al., A consensus genome-scale reconstruction of Chinese hamster ovary cell metabolism, Cell Syst. 3 (5) (2016) 434–443. |
| Contact: | M. Masid , V. Hatzimanikatis |
Comparison of the metabolism between various malaria parasites
 Malaria is a major global health care concern, and kills half a million people each year. It is caused by protozoan parasites of the genus Plasmodium. There are five parasites capable of infecting humans and their metabolism is slightly different. The eradication of malaria requires the identification of drug targets in all of the malaria parasites.
Malaria is a major global health care concern, and kills half a million people each year. It is caused by protozoan parasites of the genus Plasmodium. There are five parasites capable of infecting humans and their metabolism is slightly different. The eradication of malaria requires the identification of drug targets in all of the malaria parasites.
In this project, we aim to perform comparative analysis of the malaria parasites’ metabolism for the identification of common drug targets. We will use genome-scale metabolic models (GEMs) of the malaria parasites and apply constrained-based approaches like Flux Balance Analysis (FBA) and Thermodynamics-based Flux Analysis (TFA). These tools serve to perform mass and energy balances on the GEMs of the parasites. We will investigate in-depth the metabolic networks and identify the metabolic differences that define the characteristic behavior of each Plasmodium species.
| Type: |
Semester or master project |
| Keywords: | Cellular metabolism, genome-scale metabolic model, malaria, drug targets, Plasmodium species |
| Recommended knowledge: | Background on mass and energy balances, general background on biochemistry, MATLAB programming |
| Suggested reading: | http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005397 |
| Contact: | V. Hatzimanikatis |
Discovery and exploration of novel biotransformation networks
 Today’s knowledge of biochemistry is far from being complete, especially when it comes to the more exotic biosynthesis routes of secondary metabolites. The pathways of peripheral metabolism produce highly complex molecules, some of which have valuable pharmaceutical or industrial properties. Many of these highly interesting metabolites have been extracted from plants and characterized, but their complex biosynthesis pathways remain unknown.
Today’s knowledge of biochemistry is far from being complete, especially when it comes to the more exotic biosynthesis routes of secondary metabolites. The pathways of peripheral metabolism produce highly complex molecules, some of which have valuable pharmaceutical or industrial properties. Many of these highly interesting metabolites have been extracted from plants and characterized, but their complex biosynthesis pathways remain unknown.
Using the computational tool BNICE.ch we can predict novel biotransformations and novel metabolites, which helps us to complement existing knowledge and to expand metabolic networks towards novel, hypothetical biosynthesis routes. We further use network analysis tools in combination with biological and chemical databases to explore and analyze, and evaluate possible biosynthesis routes that produce interesting pharmaceutical compounds. The output of our analysis will shed light on the metabolic processes and, at the same time, help metabolic engineers to modify organisms for the controlled production of these chemicals.
| Type: | Semester or master project |
| Keywords: | Novel biotransformations, biosynthetic reaction networks |
| Recommended knowledge: | Background in biochemistry and/or bioinformatics |
| Suggested reading: | Hadadi, N., et al., ATLAS of Biochemistry: A Repository of All Possible Biochemical Reactions for Synthetic Biology and Metabolic Engineering Studies. Acs Synthetic Biology, 2016. 5(10): p. 1155-1166 |
| Contact: | V. Hatzimanikatis |
Classification of Reaction Kinetics in Metabolic Networks
 Stoichiometric models of large-scale metabolic networks are widely used to perform flux balance analysis, and explore different steady state characteristics of complex biochemical systems. However, these models cannot be used the study the dynamic properties of these systems, such as transitions between steady states. For this purpose it is required to take into account the information about the kinetic properties of these reaction networks. Incorporating this information for the generation of kinetic models requires the formulation of the reaction rate expressions for each and every individual biochemical reaction in the system. To build the rate expressions we need to consider the properties of each reaction regarding its molecularity, its thermodynamic and kinetic properties. Although the kinetic mechanism of many reactions is well studied, in most cases the exact kinetic mechanism is not known. Therefore, under different assumptions, distinct generalized approximations are often used.
Stoichiometric models of large-scale metabolic networks are widely used to perform flux balance analysis, and explore different steady state characteristics of complex biochemical systems. However, these models cannot be used the study the dynamic properties of these systems, such as transitions between steady states. For this purpose it is required to take into account the information about the kinetic properties of these reaction networks. Incorporating this information for the generation of kinetic models requires the formulation of the reaction rate expressions for each and every individual biochemical reaction in the system. To build the rate expressions we need to consider the properties of each reaction regarding its molecularity, its thermodynamic and kinetic properties. Although the kinetic mechanism of many reactions is well studied, in most cases the exact kinetic mechanism is not known. Therefore, under different assumptions, distinct generalized approximations are often used.
| Type: | Semester project |
| Keywords: | Reaction classification |
| Recommended knowledge: | Chemistry/Chemical engineering, Bio-Informatics |
| Suggested reading: | Heinrich, R., & Schuster, S. (2012). The regulation of cellular systems. Springer Science & Business Media. |
| Segel, I. H. (1975). Enzyme kinetics (Vol. 957). Wiley, New York. | |
| Contact: | V. Hatzimanikatis |