We use Embryonic Stem (ES) cells as a main model system to study cell identity. ES cells are derived from the inner cell mass of the embryo at the blastocyst stage. These serve as a powerful paradigm of cell identity control, as they can be maintained a self-renewing state or instructed to differentiate towards virtually any cell type of the body. In addition, they are a promising source for future cell therapy applications, which aim at replacing cells lost in pathological conditions such as Parkinson’s disease, myocardial infarction, diabetes, and other major human diseases. We are also studying pathological regulation of cell identity in the context of cancer, with a focus on self-renewal of cancer stem cells.
Our laboratory develops quantitative live cell imaging approaches of mammalian gene expression, in order to understand how gene expression is regulated at the single cell level and throughout the cell cycle of self-renewing and differentiating stem cells. We perform quantitative analysis of transcription factor concentration and activity at the single cell and single molecule level, to understand how their activity depends on their biophysical properties, temporal fluctuations of their expression levels, and interplay with the cell cycle. We have also developed several approaches allowing quantitative monitoring of transcription, protein synthesis and protein degradation rates in single living cells. We combine these live cell technologies with genome-wide analysis of transcription factor binding (ChIP-seq, CUT&RUN), chromatin accessibility (ATAC-seq) and transcription (RNA-seq).
Selected Publications
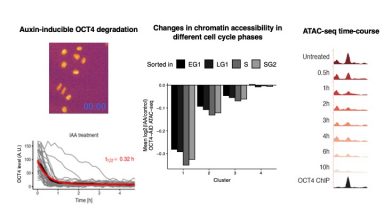
Dynamic regulation of chromatin accessibility
The two main findings of this work are: i) although mostly bound together as a heterodimer, the pioneer transcription factors OCT4 and SOX2 act in a largely independent manner to modulate chromatin accessibility; ii) OCT4 maintains chromatin accessibility in a highly dynamic manner (on a minute time-scale) throughout the cell cycle of ES cells.
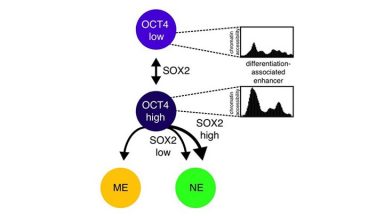
Transcription factor fluctuations bias cell fate decisions
This paper shows that mild (about 2-fold) endogenous temporal variations in the concentrations of OCT4 and SOX2 strongly bias differentiation efficiency of ES cells towards neuroectoderm and mesendoderm.
Transcriptional memory
Using ultrasensitive monitoring of transcription by luminescence microscopy, we show how variability in transcriptional activity propagates through cell lineages. We found that gene expression memory scales with transcriptional variability, in other words genes that are “noisy” at the population level display longer transcriptional memory.
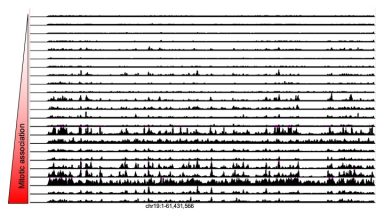
Transcription factor search
In this paper we describe our discovery that different transcription factors vary broadly (over 3 orders of magnitude) in their ability to find their specific site in the genome. Their search capability can be predicted by their non-specific DNA binding properties, which can itself be quantified by their ability to associate with mitotic chromosomes.
Proteostasis at the single cell level
We developed a method based on live imaging of a fluorescent timer fused to endogenously expressed proteins, allowing to measure protein synthesis and degradation rates simultaneously in live cells. We show how these two parameters interplay to regulate protein accumulation over the cell cycle, and that cells vary broadly in their global protein degradation rates but can compensate for this variability by adjusting their protein synthesis rates.
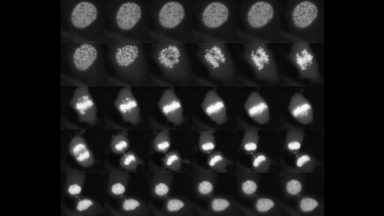
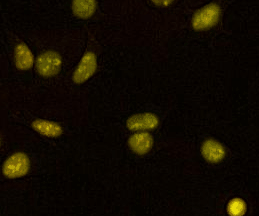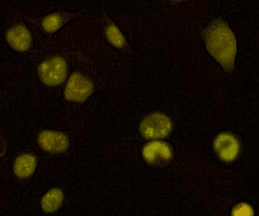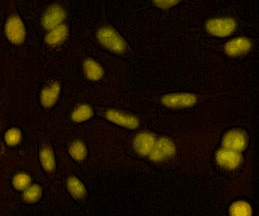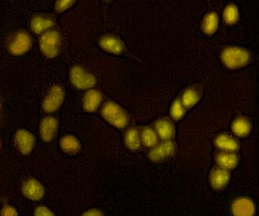
Sox2 controls cell identity during mitotic exit
Here we show that the transcription factor SOX2 associates with mitotic chromosomes, and that its depletion at the mitosis-G1 transition impairs its function in regulating self-renewal and differentiation of ES cells. This was the first demonstration of the role of a transcription factor at the mitosis-G1 transition in cell fate determination.
Earlier selected publications
Molina N*, Suter DM*†, Zoller B, Gotic I, Naef F†. Stimulus-induced modulation of transcriptional bursting in a single mammalian gene. *Equal contribution; †Corresponding authors. PNAS, 2013 Dec 17
Gebhardt JCM* Suter DM*, Roy R, Zhao ZW, Chapman A, Basu S, Maniatis T, Xie XS. Probing Transcription Factor DNA Binding at the Single Molecule Level in Live Mammalian Cells. *Equal contribution. Nature Methods, 2013 Aug;10(8):692
Stratmann M*, Suter DM*, Molina N, Naef F, Schibler U. Circadian Dbp Transcription Relies on Highly Dynamic BMAL1-CLOCK Interaction with E Boxes and Requires the Proteasome. *Equal contribution. Molecular Cell. 2012 Oct 26
Suter DM*, Molina N*, Gatfield D, Schneider K, Schibler U, Naef F. Mammalian genes are transcribed with widely different bursting kinetics. *Equal contribution. Science. 2011 Apr 22