Exploring the Beauty of Protein-Based Polymers: A Master Thesis Project
Since the inception of Bakelite in 1907, polymers have emerged as integral components of our society. However, the exponential growth of global plastic production, primarily derived from petroleum-based chemicals, has raised serious sustainability concerns. As a result, the imperative to seek environmentally friendly alternatives has become increasingly evident. Among these alternatives, natural polymer waste, especially protein waste, stands out as a highly promising candidate.
Our research group has recently developed an innovative approach known as Nature-Inspired Circular-Economy Recycling ( NaCRe ) to address the recycling of proteins once they reach the end of their lifecycle. This groundbreaking methodology involves the enzymatic depolymerization of proteins into amino acids, which are subsequently utilized in the extracellular ribosomal synthesis of new proteins. NaCRe presents a novel avenue for the efficient recycling of proteins.
However, before proteins can replace plastics on a large scale, a fundamental challenge must be addressed. Proteins, by their nature, often exhibit low toughness and limited chemical resistance due to their linear and rigid chemical structure. These inherent limitations significantly hinder the broad application of protein-based materials.
In this research project, we will employ various advanced chemical techniques (chemical synthesis, chemical modification, polymerization, material characterization, etc.) to address this challenge. Our primary objective is to enhance the mechanical properties of protein-based materials without compromising their recyclability. Simultaneously, we will explore the various potential applications of these improved materials.
We invite prospective master’s students possessing a diverse range of backgrounds, including chemistry, polymer materials, biology, and related fields. What units us is a shared and unparalleled enthusiasm for delving deep into the fascinating world of protein-based polymer materials, uncovering their inherent beauty, and advancing our understanding of their intricate properties and applications.
Contact: Dr. Youwei MA
Email: [email protected]
Glycoprotein-Based Hydrogels for Neural Regenerative Medicine Applications
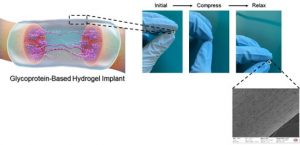
Hydrogels have high water absorption and high capability to accommodate, encapsulate, and release therapeutic components such as growth factors, drugs, and cells. Therefore, they closely mimic native biological tissues, which makes them ideal biocompatible materials for regenerative medicine/tissue engineering applications. Depending on their composition, their biological, chemical, and mechanical properties can be tuned so that the hydrogel-tissue interface has a minimal mismatch with the native tissue.
In this project, we will produce protein and carbohydrate-based bioactive hydrogels to be used as an implant to treat peripheral nerve injuries. We will investigate the effect of composition on the physicochemical, mechanical, and biological properties. We will characterize the prepared hydrogels to confirm their properties. If you are interested in biomaterials and want to broaden your knowledge/experience in chemistry and biology, please get in touch with us!
Contact: Melis Özkan, MXH130, [email protected]
Modulation of protein-ligand binding by small molecules
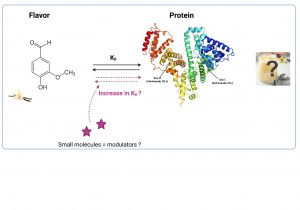
Flavor molecules in food interact with different ingredients, and this has been reported to decrease the perception of these flavor molecules. One of the most important reactions is the interaction between flavors and proteins. The objective of the project is to determine if and how small molecules modulate flavor-protein interactions in order to enhance flavor perception. In order to achieve such a goal, studies first in solutions (model systems) need to be performed to better understand and control flavor-protein binding.
The proposed semester project has an objective to characterize and better understand how specific and known small molecules can modulate the binding of ligands (eg, flavor molecules) to proteins in solution. The final goal is to be able to unravel the mechanism of such molecules. The project stands between biochemistry and solution science. Several techniques can be used to better understand the role of small molecules in protein solutions, such as osmometry, absorbance/fluorescence, and Isothermal Titration Calorimetry (ITC). This project is a good opportunity to better understand the protein state in solution and be familiarized with common and more expert characterization techniques.
No specific background is required.
Contact: [email protected]
Tailoring Lipid Nanoparticle Chemistry for Nucleic Acid Drug Delivery
Lipid nanoparticles (LNPs) have emerged as promising delivery vehicles for nucleic acid drugs for therapeutics and vaccines. The chemical nature of LNPs, for example, lipid chemical structure, lipid composition, and surface ligands may affect the delivery efficiency of nucleic acid cargo. In this project, we will seek to understand how chemically diverse LNPs differ in their physical properties (size, charge, encapsulation efficiency, etc.) and biological behaviors (transfection efficiency, endocytosis, etc.). You will:
(1) Synthesize LNPs composed of various lipid components with a microfluidic device and characterize their size distribution, zeta potential, and encapsulation efficiency of RNA drugs.
(2) Explore the possibility of introducing surface modifications and tailoring surface properties of LNPs.
(3) Evaluate intracellular delivery efficiency by in vitro experiments.
References:
1. Dilliard, SA &vSiegwart, DJ Passive, active and endogenous organ-targeted lipid and polymer nanoparticles for delivery of genetic drugs. Nat. Rev. Mater. 8, 282–300m (2023).
2. Paunovska, K., Loughrey, D. & Dahlman, JE Drug delivery systems for RNA therapeutics. Nat. Rev. Broom. 23, 265–280 (2022).
3. Lokugamage, MP et al. Optimization of lipid nanoparticles for the delivery of nebulized therapeutic mRNA to the lungs. Nat. Biomed. Eng. 5, 1059–1068 (2021).
4. Hatit, MZC et al. Species-dependent in vivo mRNA delivery and cellular responses to nanoparticles. Nat. Nanotechnology. 17, 310–318 (2022).
5. Dobrowolski, C. et al. Nanoparticle single-cell multiomic readouts reveal that cell heterogeneity influences lipid nanoparticle-mediated messenger RNA delivery. Nat.Nanotechnol. 17, 871–879 (2022).
6. Dilliard, SA, Cheng, Q. & Siegwart, DJ On the mechanism of tissue-specific mRNA delivery by selective organ targeting nanoparticles. proc. Natl. Acad. Sci. USA 118, (2021).
7. Cheng, Q. et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnology. 15, 313–320 (2020).
8. Melamed, JR et al. Lipid nanoparticle chemistry determines how nucleoside base modifications alter mRNA delivery. J.Control. Release 341, 206–214 (2022).
Contact: Heyun Wang ([email protected])
How to quantify protein interactions in the presence of small molecules?
Weak interactions are predominantly responsible for proteins to form complex colloids and solutions. However, they are not fully understood yet. When studying solutions, weak interactions and their binding properties are quantified using the osmotic second virial coefficient and the equilibrium dissociation constant. Recently, Self-Interaction Chromatography (SIC) has been introduced as an efficient technique and with the unique capability to be extended to Cross-Interaction Chromatography (CIC) quantifying the interaction between two different proteins. To measure the binding affinities and kinetics of proteins and protein-ligands with high-throughput and high sensitivity, the Surface Plasmon Resonance (SPR) technique has yielded rapid and reliable results across fields and industries ranging from pharmaceutical to food sciences. We successfully combine SIC/CIC and SPR experiments to quantify weak interactions between proteins and protein isolates in different solution conditions. Furthermore, we perform more standard measurements on the proteins of interest in different buffer conditions and in the presence of small molecules. We conduct absorbance and static light scattering measurements to determine the size and the presence of large aggregates, circular dichroism measurements to determine the secondary structure, and we evaluate the respective zeta potential of the protein of interest.
However, there are still many phenomena that we do not fully understand, and so we are continuing to explore different proteins the effect of a broad range of small molecules, and we are also adding more techniques to our toolset. Our goal is to understand and describe the driving mechanism behind the effect of small molecules on protein interactions. Thus, in case you are interested and or have more questions, please get in touch!
Here is a small teaser which includes a tasty potential application:
https://drive.google.com/file/d/1qOl78vni55OC9bLqFPIxyEKMVOHnjonx/view?usp=share_link
Contact: Pamina Winkler (email: [email protected])
The effect of amino acids mixtures on colloidal stability of protein systems
The colloidal stability of proteins, which refers to their ability to remain in solution without aggregating or precipitating out of solution, is essential for many pharmaceutical, food, and biotechnological applications. For example, colloidal aggregation of proteins can affect their efficacy and safety as therapeutic agents and can also lead to various diseases, including blood coagulation disorder, cataracts, and atherosclerosis. Therefore, understanding the factors affecting the aggregative stability of protein dispersions is crucial for drug development, disease treatment and prevention, and advancing our fundamental knowledge of these biological systems.
The cellular environment is rich in small molecules, such as amino acids, influencing protein-protein interactions and affecting protein aggregation stability. Some amino acids and their mixtures have been widely used for protein stabilization in medical formulations. However, the mechanism behind the stabilizing effect of amino acids has yet to be fully studied. Furthermore, understanding whether the effect of amino acid mixtures is additive, synergistic, or antagonistic could lead to the development of more effective protein stabilization techniques.
In this project, quantitative evaluation of the colloidal stability of protein systems will be done by studying liquid-liquid phase separation (LLPS) – a process in which proteins transition from a homogeneous solution to two coexisting liquid phases. The research will focus on determining the amino acid mixture’s effect on protein LLPS and identifying the underlying mechanisms that drive these effects.
If you are interested in this project, please do not hesitate to contact us.
Contact: Ekaterina Poliukhina, MXH130 Email: [email protected]