Segmentation Timing and Dynamics Laboratory
How do spatio-temporal patterns emerge at the tissue level from noisy cellular and molecular interactions? What are the principles that govern transitions from parts to wholes, and those that determine precision and robustness? We explore these issues using a population of genetic oscillators in the vertebrate embryo termed the segmentation clock.
This multi-cellular clock drives the rhythmic, sequential, and precise formation of embryonic body segments, exhibiting rich spatial and temporal phenomena spanning from molecular to tissue scales. Tissue patterning by cellular oscillations is a recent concept, and the mechanisms and molecules responsible for this astonishing activity are just beginning to be understood.
We are biologists, engineers and physicists using molecular genetics, quantitative imaging and theoretical analysis. Because timing is the key to understanding oscillations, we developed a multiple-embryo time-lapse recording method to enable the quantitation and statistical treatment of somitogenesis dynamics. To understand the interactions of noisy cellular oscillators and regulatory networks, we developed theoretical descriptions that have been successfully tested in embryos. We identified the first mutations that change the period of the segmentation clock, opening the door to a molecular understanding of the mechanisms that control the clock’s dynamics. We are beginning to explore new transgenic tools that allow us to follow the oscillations of the segmentation clock in real time. One of these revealed the existence of a Doppler effect associated with the wave patterns of gene expression and the shrinking tissue length in the embryo.
The vertebrate segmentation clock, a rhythmic, tissue-level pattern generator responsible for sequential formation of the embryonic segments that give rise to backbone, ribs and associated muscles. For more details, see: Oates AC, Morelli LG, Ares S. Patterning embryos with oscillations: structure, function, and dynamics of the vertebrate segmentation clock. Development. 2012 Feb 139:625-639.
Current third party funding Wellcome Trust Senior Research Fellowship in Basic Biomedical Science (WT098025MA)
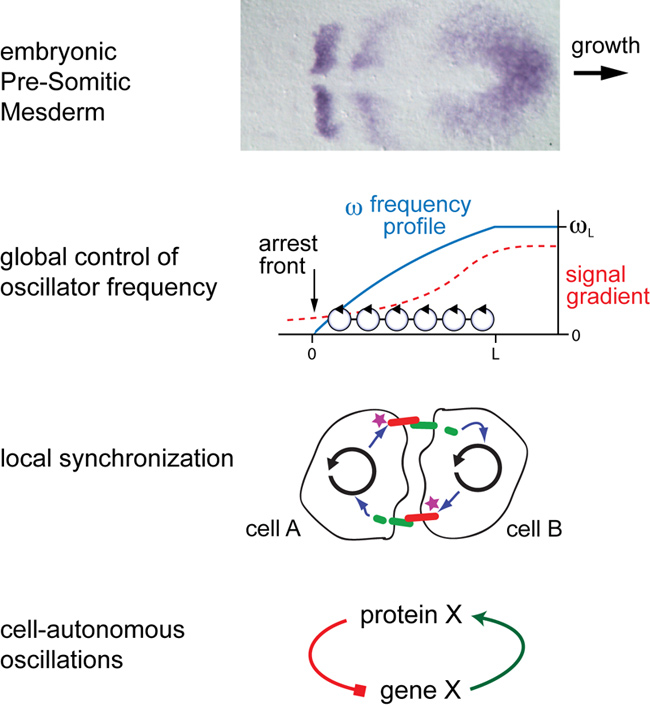
Projects (theory and experiment) – Single cell oscillations – Cell-cell synchronization – Global control of oscillator arrest – Cellular movement and segmentation
Collaborators – Professor Frank Jülicher, Max Planck Institute for the Physics of Complex Systems, Dresden, Germany – Dr Luis Morelli, Departamento de Física, Universidad de Buenos Aires, Buenos Aires, Argentina – Dr Saul Ares, Logic of Genomic Systems Laboratory, Centro Nacional de Biotecnología – CSIC, Madrid, Spain – Dr Koichiro Uriu, Theoretical Biology Laboratory, RIKEN Advanced Science Institute, Saitama, Japan
Selected publications
Rohde LA, Bercowsky-Rama A, Negrete J Jr, Valentin G, Naganathan SR, Desai RA, Strnad P, Soroldoni D, Jülicher F, Oates AC. Cell-autonomous generation of the wave pattern within the vertebrate segmentation clock. BioRxiv. May 30, 2021. Doi https://doi.org/10.1101/2021.05.29.446196.
Fantner GE, Oates AC. Instruments of change for academic tool development. 2021. Nature Physics, Volume 17, Issue 4, p.421-424.
Negrete J Jr, Oates AC. Towards a physical understanding of developmental patterning. Nat Rev Genet. 2021 May 10. Doi: 10.1038/s41576-021-00355-7. Online ahead of print. PMID: 33972772 Review.
Uriu K, Liao BK, Oates AC, Morelli LG. From local resynchronization to global pattern recovery in the zebrafish segmentation clock. Elife. 2021 Feb 15;10:e61358. doi: 10.7554/eLife.61358. PMID: 33587039 Free PMC article.
Negrete J Jr, Lengyel IM, Rohde LA, Desai RA, Oates AC, Jülicher F, Theory of time delayed genetic oscillations with external noisy regulation, New Journal of Physics, 2021.
Naganathan SR, Oates AC. Patterning and mechanics of somite boundaries in zebrafish embryos, Seminars in Cell & Developmental Biology, Elsevier, November 2020, Volume 107, 170-178.
Naganathan SR, Popovic M, Oates AC. Somite deformations buffer imprecise segment lengths to ensure left-right symmetry. BioRxiv. August 14, 2020.
doi. https://doi.org/10.1101/2020.08.14.251645.
Oates AC. Waiting on the Fringe: cell autonomy and signaling delays in segmentation clocks. Curr Opin Genet Dev. 2020 Aug;63:61-70. doi: 10.1016/j.gde.2020.04.008. Epub 2020 Jun 5. PMID: 32505051 Free article. Review.
Venzin OF, Oates AC. What are you synching about? Emerging complexity of Notch signaling in the segmentation clock. Dev Biol. 2020 Apr 1, 460(1):40-54.
Narayanan R, Oates AC. Detection of mRNA by Whole Mount in situ Hybridization and DNA Extraction for Genotyping of Zebrafish Embryos Bio-Protocol. 2019. 9 (6), e3193.
Negrete J Jr, Oates AC. Embryonic lateral inhibition as optical modes: An analytical framework for mesoscopic pattern formation. Physical Review E. 2019. 99 (4), 042417.
Lleras Forero L, Narayanan R, Huitema LF, VanBergen M, Apschner A, Peterson-Maduro J, Logister I, Valentin G, Morelli LG, Oates AC, Schulte-Merker S. Segmentation of the zebrafish axial skeleton relies on notochord sheath cells and not on the segmentation clock. Elife. 2018 Apr 6;7.
Richter S, Schulze U, Tomançak P, Oates AC. Small molecule screen in embryonic zebrafish using modular variations to target segmentation. Nat Commun. 2017 Dec. 1;8(1):1901.
Uriu K, Bhavna R, Oates AC, Morelli LG. A framework for quantification and physical modeling of cell mixing applied to oscillator synchronization in vertebrate somitogenesis. Biol Open. 2017 Aug. 15;6(8):1235-1244.
Liao BK, Oates AC. Delta-Notch signalling in segmentation. Arthropod Struct Dev. 2017 May. 46(3):429-447.
Naganathan SR, Oates AC. The Sweetness of Embryonic Elongation and Differentiation. Developmental Cell. 2017 Feb 27, 40(4):323-324.
Jörg DJ, Oates AC, Jülicher F. Sequential Pattern Formation Governed by Signaling Gradients. Physical Biology. 2016 13, 05LT03.
Liao BK, Jörg DJ, Oates AC. Faster embryonic segmentation through elevated Delta-Notch signalling. Nature Communications. 2016 Jun 15;7:11861.
Rajasekaran B, Uriu K, Valentin G, Tinevez JY, Oates AC. Object Segmentation and Ground Truth in 3D Embryonic Imaging. PLoS One 2016 Jun 22;11(6): e0150853.
Webb AB, Lengyel IM, Jörg DJ, Valentin G, Jülicher F, Morelli LG, Oates AC. Persistence, period and precision of autonomous cellular oscillators from the zebrafish segmentation clock. Elife. 2016 Feb 16;5. pii: e08438.