Prof. La Manno Lab – Laboratory of Brain Development and Biological Data Science
A significant aspect of our research is the exploration of lipid heterogeneity at the single-cell level. We believe this emerging field holds the key to understanding how different biochemical drivers, including lipids, influence the dynamic process of brain development.
Our long-term goal is to unravel the intricate gene regulatory programs that drive the formation of cell types in the nervous system. By doing so, we hope to contribute to the understanding of neurodevelopmental disorders and the improvement of cell replacement therapies.
Selected Publications
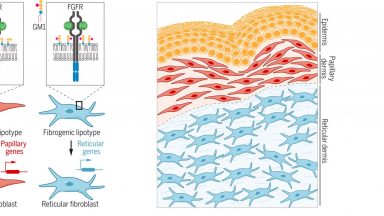
Sphingolipids Control Dermal Fibroblast Heterogeneity
Cell-to-cell lipid heterogeneity affects the determination of cell states, adding a new regulatory component to the self-organization of multicellular systems.
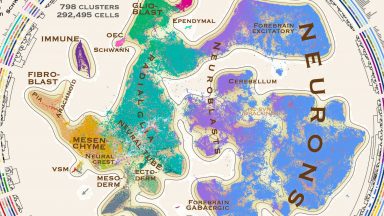
Molecular architecture of the developing mouse brain
We report a comprehensive single-cell transcriptomic atlas of the embryonic mouse brain between gastrulation and birth. We identified almost eight hundred cellular states that describe a developmental program for the functional elements of the brain and its enclosing membranes.
Spatial tissue profiling by imaging-free molecular tomography
We describe an imaging-free framework to localize high-throughput readouts within a tissue by cutting the sample into thin strips in a way that allows subsequent image reconstruction. We applied the technique to the brain of the Australian bearded dragon, Pogona vitticeps. Our results reveal the molecular anatomy of the telencephalon of this lizard, providing evidence for a marked regionalization of the reptilian pallium and subpallium.
LATEST PUBLICATIONS
Clinically compliant cryopreservation of differentiated retinal pigment epithelial cells
Cytotherapy. 2024-04-01. Vol. 26, num. 4, p. 340-350. DOI : 10.1016/j.jcyt.2024.01.014.Deciphering the nature of cell state transitions in single cells using quantitative modeling of temporal dynamics
Lausanne, EPFL, 2024.Meeting our Makers: Uncovering the cis-regulatory activity of transposable elements using statistical learning
Lausanne, EPFL, 2024.Contact
Head of Laboratory : Gioele La Manno
Office : AAB 241
Phone : +41 21 69 38446
Mail : [email protected]
Administrative Assistant : Silvia Avanzi