Contact:
Abhishek Garg, PhD Student, EPFL-IC-LSI
Partners:
Ioannis Xenarios, Merck-Serono, Geneva, Switzerland
Keywords: gene regulatory networks, signalling pathways, boolean networks, BDDs, microarrays
Presentation:
The face of biological research has evolved at an alarming rate over the last two decades. From a one-gene/one-protein analysis it has borne witness to a multitude of technologies that allows us to capture and integrate a vast amount of information generated by high throughput methods such as DNA microarrays, siRNA knock-down and protein-protein interactions. While a wealth of information is present on the interaction of the genes and proteins, the exact stoichiometry and precise kinetics still evades our technologies and understanding. In such situation, one could either wait to gather the crucial information on the precise biochemical processes or choose to model the flow of information in genetic regulatory networks. In this project we chose the latter as we think that the information is sufficient already to identify qualitative behavior of the studied biological system. We also believe that enabling such kind of approaches should further the understanding of the design and identifications of keys elements that dictate cell fate.
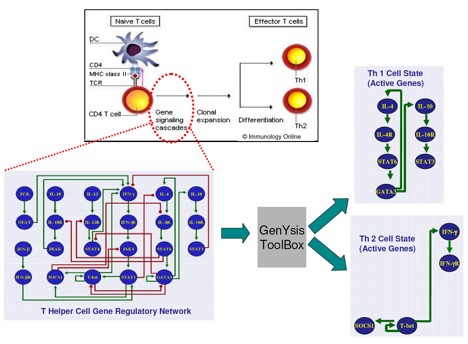
An application of GenYsis to find cell states of T Helper Cell
Goal:
The goal of the project is to develop algorithms for reconstructing GRN from microarray data and methods for performing in silico experiments on these networks. The techniques developed are at the interface of knowledge from the fields of Electronic Design Automation, Model Checking and Verification, and Machine Learning.
We have developed a tool called GenYsis for modeling dynamics of gene regulatory networks using Binary Decision Diagrams (BDDs). GenYsis can find steady states of the regulatory networks. These steady states reflect the fate of the cell (corresponding to the network). In addition genYsis can perform in silico gene perturbation experiments on these networks. Such experiments can help biologists to understand the cell behavior and they can use this tool in more advanced cases for modeling the immune systems.
GenYsis is being constantly extended to add new functionalities. Currently we are extending this tool for automatic reconstruction of Gene Regulatory networks from high throughput data like micro arrays.
Publications:
Modeling of Multipled Valued Gene Regulatory Networks, Abhishek Garg, Luis Mendoza, Ioannis Xenarios and Giovanni DeMicheli, 29th IEEE EMBS Annual International Conference 2007, Lyon, France (to appear).
An Efficient Method for Dynamic Analysis of Gene Regulatory Networks and in-silico Gene Perturbation Experiments , Abhishek Garg, Ioannis Xenarios, Luis Mendoza, Giovanni DeMicheli, 11th Annual International Conference on Research in Computational Molecular Biology 2007. Published in Springer’s Lecture Notes in Computer Science, Volume 4453.
Exploiting Binary Abstractions in Deciphering Gene Interactions , Sungroh Yoon, Abhishek Garg, Eui-Young Chung, Hyun Seok Park, Woong Yang Park, Giovanni De Micheli. , 28th IEEE EMBS Annual International Conference 2006, New York, USA.
Download project description file (578 KB pdf)