Our laboratory is interested in two large research avenues. The first is concerned with our historical interest in the study of Hox gene regulation during mammalian development. The main projects are mentioned below, under the research plan of a recently funded grant application. Long-range gene regulation at Hox loci is studied by using CRISPR-cas9 genomic editing both in mice and in ES-cells derived pseudo-embryos (gastruloids), which are excellent paradigms to look at the molecular basis of body axis extension in mammals. For the past three years, a strong effort was implemented to reallocate forces towards this model system, for it allows a high precision in developmental staging, an easy access to large amounts of material and to its visualization and the possibility to mutate ES cells instead of mouse zygotes.
Our second line of research is part of a common effort between our laboratory and that of Prisca Liberali (FMI, Basel) and Mathias Lutolf (EPFL, Lausanne), funded by a collaborative grant, to look at the effects of early mechanical constrains on the development of gastruloids, in particular at the start of axial extension. Our main projects along this line are also mentioned below, as part of another funded grant application.
1) GENE REGULATION AT HOX LOCI DURING AXIAL EXTENSION IN VIVO AND IN VITRO
The Hox genes family encode series of transcription factors with a key function in organizing various aspects of the developing bodies of all animals displaying a bilateral symmetry, including vertebrates and hence mammals. Ever since their discovery in the mid 1980’s, both their function and their transcriptional regulation have been heavily studied in many different systems, and critical concepts have derived from these studies, which has made this gene family a spectacular epistemic system. However, due to their intrinsic properties, two of the most essential questions have remained unanswered. The first one has to do with the mechanism underlying the time sequence of Hox genes transcriptional activation following their physical order along the different clusters. Thus far, the understanding of this fundamental mechanism has resisted many years of experimental approaches.
The second question is that of the basic function of this gene family when taken as a whole rather than as a collection of separate elements. Indeed, while we know quite well the function of each Hox gene individually, we do not know what would be the effect on embryo development of removing either large parts of- or all of this gene family. The reasons for this failure are numerous but mostly based on the experimental difficulties to study a complex mechanism acting right at the worst development stage to work with the mouse embryo. The accessibility of the material, the small number of cells concerned, the heterogeneity in the ages of littermates as well as all kind of experimental hurdles have thus hampered the solutions to these questions. Recently, various systems were developed which allow for pseudo-embryos to be produced in vitro starting exclusively from ES cells, which can thus be easily modifiable through CRISPR-Cas9 approaches. We thus believe that these two major questions can now be answered by using this alternative system.
Overall objectives: The two overarching aims of this application are 1) to decipher the mechanism implementing the time-sequenced transcriptional activation of Hox genes along their respective clusters (the ‘Hox clock’) and 2) to reveal the deep functional importance of HOX proteins during the development of vertebrates. We believe these are two main fundamental questions left to be answered concerning the Hox gene family and we trust that a drastic switch in our experimental model and tools may help us to eventually achieve this task.
Specific aims: For overarching aim 1, we shall first calibrate gastruloids (pseudo-embryos extending a ‘body axis’, see below) to precisely describe the Hox clock in these biological objects (a clock, which we know is perfectly implemented). Once this will be done, we will start a series of mutations in ES cells, either affecting the genomic structure of Hox clusters or modifying (adding, deleting) CTCF binding sites and produce mutant gastruloids, which will be analyzed using a pipeline of readouts (RNA-seq, ATAC-seq and ChIP-seq or Cut&Run of PolII, Rad21, CTCF, Nipbl as well as a variety of histone H3 tail modifications). In this experimental configuration, even minor changes in timing will be scored. For overarching aim 2, we shall produce series of ES cell lines carrying massive variations in their number of Hox gene and clusters. Two specific aims will be targeted, the first being the functional removal of entire paralogy groups (i.e. all Hox gene derived from ancestral duplication events), in particular all Hox1 and all Hox13 genes. The second will aim at removing the entire Hox gene family (i.e. 4 ca. 100 kb large deletions on different chromosomes) to assess whether Hox function is only required to identify body parts (as in invertebrates) or if is it also involved in the production thereof, as is the case during vertebrate limb development. The experimental readout will consist of single cells RNA-seq, which should give us a molecular readout of the perturbations, as gastruloids do not develop full embryonic structures.
Research Plan
Research projects
|
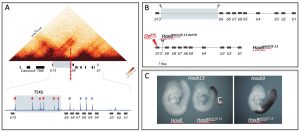
AIM 1. The impact of chromatin architecture on Hox gene activation in space and time 1.3.1. HoxD CTCF Mutagenesis in vivo 1.3.1.1. Effects on the timing of Hox gene activation 1.3.1.2. Effects on the spatial distribution of Hox gene transcript domains 1.3.2. Mutagenesis of the HoxB TAD boundary 1.3.3. HoxD mutagenesis in gastruloids in vitro 1.3.3.1. Production of Hox clusters and Hox loci haploid cell lines and Hox-GFP cell lines 1.3.3.2. CTCF Mutations and deletions at HoxD. Effects on timing and opening 1.3.3.3. Boundary deletions and reconstitution at HoxB CTCF mutations AIM 2. Assessing Hox gene function in gastruloids 1.3.4. Function of Hox genes during gastruloid extension 1.3.4.1. Single-cell RNA-seq as a phenotypic tool. 1.3.4.2. Function of anterior Hox genes during trunk extension 1.3.4.3. Multiple loss of functions 1.3.5. Function of Hox13 genes in the termination of the body axis 1.3.5.1. Combined KO of group 13 genes 1.3.5.2. Effect of Hoxb13 gain of function during axial extension 1.3.6. Potential avenues to explore 1.3.6.1. iPS-derived gastruloids from mutant stocks 1.3.6.2. Elongation of gastruloids in the complete absence of Hox functions. 1.3.6.3. Final validation in embryos through tetraploid chimaeras. |
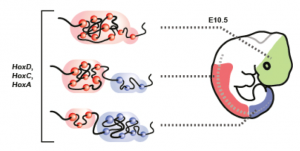
Figure 1. The rosary model, shown as a fixed time point (ca. 11.5 old embryo. In the developing brain (green), the Hox cluster is compacted as a negative chromatin domain (in pink), covered by polycomb-associated marks (Vieux-Rochas et al., 2015). In the anterior trunk (red), some genes have been ‘opened’ (blue balls) and are now in a ‘positive’ chromatin domain, labelled by H3K4me3. In more posterior cells (right, blue), more genes have been opened until the last group 13 genes become transcriptionally competent. The states represented in the left illustrate memorized states produced in time alng with the extension of the body axis (from Noordermeer et al., 2014). |
2) ENGINEERING GASTRULOIDS TO STUDY THE IMPACT OF MECHANICAL CONSTRAINTS UPON MAMMALIAN DEVELOPMENT
The impact of the physical environment upon the development of the mammalian embryo has been difficult to evaluate, due to the challenge to access and perturb these embryos in their native niches, at the precise time when the major decisions are taken regarding the general body architecture and the onset of cellular differentiation. In the past five years, several systems have been developed, which can recapitulate some aspects of mammalian development under in vitro conditions, amongst which gastruloids i.e. ‘pseudo-embryos’ lacking a head produced out of embryonic stem (ES) cell aggregates that can self-organize into the three major body axes. The overarching goal of our research program is to use such gastruloids to study the potential impacts of external mechanical stresses and geometrical constraints on their symmetry breaking, self-organization and axial extension. To achieve this goal, we want to set up novel analytical tools and platforms such that both normal and genetically modified gastruloids can be grown and monitored live under a variety of mechano-geometrical environmental conditions. The effects of these extrinsic physical cues upon the implementation of developmental pathways will be analyzed at the single cell level both by using fluorescent marker genes and by detailed epigenetic and transcriptomics analyses of the affected gastruloids. The results obtained will identify candidate genes or pathways potentially involved in the response of mammalian embryos to their physical environment, which will be further studied in an iterative manner, after genetic modifications, using the same approaches.
Because of both their high derivation efficiency (above 75%) and their easiness to produce under laboratory conditions, gastruloids are the only type of ‘pseudo-embryos’ that can reasonably be used in such a high-throughput experimental design. Consequently, we believe that this complementary program will deliver critical results and information regarding an aspect of mammalian development that is only starting to emerge now. We also believe that the conclusions obtained by studying the effects of physical effectors on the developing embryo will likely reflect fundamental ways that the organism implements to deal with its immediate environment and thus be applicable to a wide variety of situations occurring either in later stages of development or in homeostatic and pathological conditions. These insights should be useful even outside the fields of developmental and stem cell biology, for example in the implementation of biology inspired approaches for tissue engineering and regenerative medicine.
Research plan
|
AIM 1. Diversity of the gastruloid transcriptomes. AIM 2. Identification of marker genes. AIM 3. Chromatin dynamics in early staged gastruloids. AIM 4. Responses of gastruloids to mechanical constraints. AIM 5. Functional analyses of potential target genes or pathways. AIM 6. Multiple scRNA sequencing, local perturbations and local detection. |
Compliance to- and dissemination of- the 3R principle.
The engineering and use of ‘pseudo-embryos’ in vitro by starting from ES cells (and/or other cell types) is fully compliant with the 3R principle in animal research (Reduced, Refine and Replace). While we want to make it clear that the use of gastruloids is not aimed at replacing the use of animals in research, we also like to show that some targeted questions amongst the most-timely in the field of mammalian developmental genetics may be addressed by using such surrogate systems, often in an even more efficient manner, thus leading to an important reduction in the number of animals used either as a source of material, or as mutant stocks. It is nevertheless obvious that the final demonstration of the potential findings derived from this research will likely call for the use of genuine animals, at least still for the next few years to come. In this context, we would like to offer to everyone who likes to try and introduce such alternative approaches in her/his laboratory the possibility to easily find the competences, material and protocols such as to start rapidly and efficiently.