SEMESTER PROJECTS/LAB IMMERSIONS 2024-2025
Personalized endocrine prevention of hormone receptor-positive breast cancer progression
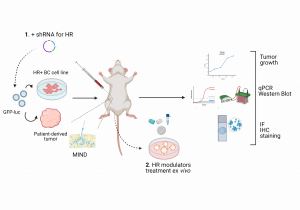
The majority of breast cancer (BC) cases are hormone receptor-positive (HR+). It is well established that estrogen receptor α (ERα) is a key driver of proliferation in HR+ BC. Nowadays, patients diagnosed with this type of BC are uniformly treated with drugs that inhibit only ERα. However, endocrine resistance and tumor recurrence remain frequent clinical problems, and new therapeutic strategies are needed. Besides ERα, such tumors also express the receptors for progesterone and testosterone, but their role in breast cancer development remains controversial and is currently not used for therapy. Using the mouse intraductal model (MIND), a physiologically relevant in vivo model to study BC, we have preliminary evidence that estrogen (ER), progesterone (PR), and androgen (AR) receptors signaling are deregulated in different ways during breast carcinogenesis in individual patients. That can provide targets for personalized prevention of BC progression, meaning that targeting PR or AR instead of ERα could benefit some patients. To study the signaling of different hormone receptors and to identify new possible targets, the student will use genetic (shRNA) and pharmacological approaches (HR modulators) in in vitro and ex vivo models of different BC cell lines and patient-derived samples. During this project, the student will master the basics of the cell culture approach, molecular biology methods, and immunohistochemistry techniques.
References:
- Sflomos, G., Dormoy, V., Metsalu, T., Jeitziner, R., Battista, L., Scabia, V., Raffoul, W., Delaloye, J.-F., et al. (2016) ‘A Preclinical Model for ERα-Positive Breast Cancer Points to the Epithelial Microenvironment as Determinant of Luminal Phenotype and Hormone Response’, Cancer Cell. Cell Press, 29(3), pp. 407–422. doi: 10.1016/j.ccell.2016.02.002. PMID: 26947176.
- Scabia, V., Ayyanan, A., De Martino, F. et al. Estrogen receptor positive breast cancers have patient specific hormone sensitivities and rely on progesterone receptor. Nat Commun 13, 3127 (2022). https://doi.org/10.1038/s41467-022-30898-0
Interested students are welcome to contact : daria.matvienko@epfl.ch
Regulation of metastatic dormancy in hormone receptor-positive breast cancer
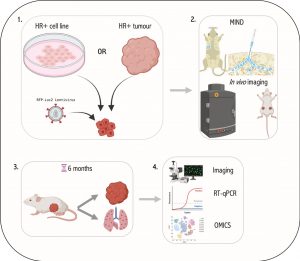
Breast cancers (BC) are divided into 4 main subtypes based on their expression of the oestrogen receptor (ER), progesterone receptor (PR) and the Human epidermal growth factor receptor (HER2) (1). ER+ breast cancer patients’ probability of survival decreases 5 – 20 years post-diagnosis due to minimal residual disease and metastatic relapse. This deadly phenomenon is caused by dormant DCCs at metastatic sites that remain undetectable at the time of initial diagnosis until giving rise to overt metastatic disease (2,3). Understanding the kinetics and molecular programmes employed by these DCCs during disease progression is essential for early intervention and prevention of this metastatic latency. In line with metastatic latency in ER+ breast cancer, it has been shown that metastatic cells from low burden tissues (HR+) displayed high expression of epithelial-to-mesenchymal transition (EMT), stem cell and dormancy genes compared to that of high burden tissues (HR-) (4,5). This suggests that cancer cells may undergo EMT and disseminate at early stages of tumour formation and seed as quiescent/dormant cells at metastatic sites for years before initiating secondary disease (6,7). As HR+ breast cancers make up roughly 70% of all breast cancers, late metastatic disease and cancer cell dormancy remains a significant clinical problem that has largely been overlooked by breast cancer researchers. This project aims to identify global regulators of EMT and metastatic dormancy in HR+ breast cancers using the MIND-model.
The student will gain experience with in vitro cell culture, ex vivo models and molecular biology techniques to tackle these questions.
References:
- Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70.
- Gawrzak S, Rinaldi L, Gregorio S, Arenas EJ, Salvador F, Urosevic J, et al. MSK1 regulates luminal cell differentiation and metastatic dormancy in ER + breast cancer. Nat Cell Biol [Internet]. 2018;20(2):211–21. Available from: http://dx.doi.org/10.1038/s41556-017-0021-z
- Zhang XHF, Giuliano M, Trivedi M V., Schiff R, Kent Osborne C. Metastasis dormancy in estrogen receptor-positive breast cancer. Clin Cancer Res. 2013;19(23):6389–97.
- Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526(7571):131–5.
- Aouad P, Zhang Y, Stibolt C, Mani SA, Sflomos G, Brisken C. Epithelial-mesenchymal plasticity determines estrogen receptor positive (ER+) breast cancer dormancy and reacquisition of an epithelial state drives awakening. bioRxiv [Internet]. 2021 Jan1;2021.07.22.453458. Available from: http://biorxiv.org/content/early/2021/07/26/2021.07.22.453458.abstract
- Hosseini H, Obradovic MMS, Hoffmann M, Harper KL, Sosa MS, Werner-Klein M, et al. Early dissemination seeds metastasis in breast cancer. Nature [Internet]. 2016;540(7634):552–8. Available from: http://dx.doi.org/10.1038/nature20785
- Jiang J, Zheng M, Zhang M, Yang X, Li L, Wang S-S, et al. PRRX1 Regulates Cellular Phenotype Plasticity and Dormancy of Head and Neck Squamous Cell Carcinoma Through miR-642b-3p. Neoplasia. 2019 Feb;21(2):216–29.
Interested students are welcome to contact : [email protected]
Suggested Projects
1. Rethinking of Hormonal Therapies in ILC Progression
Invasive lobular carcinoma (ILC) is the most common breast cancer ‘‘special type’’ and is mainly driven by the loss of E-cadherin expression. ILC responds poorly to chemotherapy and tamoxifen and is the histological subtype strongly associated with ovarian hormone usage, especially estrogens e.g., Ethinylestradiol (EE) and progestogens (e.g., medroxyprogesterone acetate). Since it has been impossible to establish xenografts from ILC cell lines and patient samples, the mechanisms underlying the characteristic biology have been challenging to address. We have recently shown that the microenvironment is critical for ER+ tumor engraftment, which is enabled by injection into the mouse milk ducts, including ILCs. We aim to address the latter biological feature of ILC by assessing the effect of different ovarian hormones in ILC breast cancer cell lines and patients’ cells in vivo and in ex vivo models of ILC. The student will master basic and advanced molecular biology methods and become familiar with in vivo and ex vivo analysis.
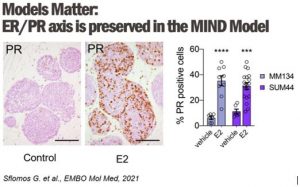
2. Dissecting the Molecular Mechanisms of Endocrine Therapy Resistance
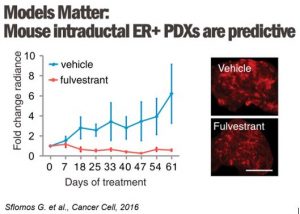
In clinics, endocrine resistance is increasingly observed following treatment with anti-estrogen therapies and aromatase inhibitors (AI). Using the intraductal transplantation method for ER+ breast cancers, we will model the clinical scenario by treating mice engrafted intraductally with either ER-positive cells MCF7 and ILC MDA-MB-134 and SUM44 with anti-estrogen therapies (tamoxifen, fulvestrant, and estrogen deprivation) to model the responses of invasive breast cancer of no special type and invasive lobular carcinomas, respectively.
We will perform transcriptomics, proteomics, and genomics analyses of the residual tumors and compare them with naïve treated xenografted tumors. We will perform endocrine treatment studies in patient-derived xenografts generated from freshly isolated breast tumors to confirm our findings. The student will master basic and advanced molecular biology methods and become familiar with in vivo and ex vivo analysis.
References
- PMID: 34771558 – https://pubmed.ncbi.nlm.nih.gov/34771558/
- PMID: 17081916 – https://pubmed.ncbi.nlm.nih.gov/17081916/
- PMID: 32508307 – https://pubmed.ncbi.nlm.nih.gov/32508307/
- PMID: 29563191 – https://pubmed.ncbi.nlm.nih.gov/29563191/
- PMID: 26977876 – https://pubmed.ncbi.nlm.nih.gov/26977876/
- PMID: 26947176 – https://pubmed.ncbi.nlm.nih.gov/26947176/
- PMID: 26241069 – https://pubmed.ncbi.nlm.nih.gov/26241069/
Interested students are welcome to contact : [email protected]
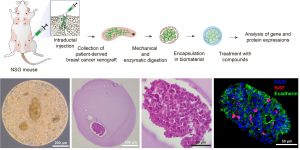
Optimizing an ex vivo assay for testing hormone and drug response in hormone-sensitive breast cancer
Breast cancer is a leading cause of cancer-related mortality for women worldwide (1). Endocrine therapy is of significant therapeutic value for hormone receptor-positive breast tumors that represent 70% of breast cancer cases (2). However, current standard endocrine therapies are largely based on pathological breast cancer features, and the outcomes are consequently affected by patient-to-patient heterogeneity. To provide tailored endocrine therapy, we propose a biomaterial-based 3D system to test hormone and drug sensitivity of patient-derived xenografts and patient tumor biopsies. The student will learn to manipulate and encapsulate tumors in biomaterials, and assess gene and protein expression using molecular biology and immunohistochemistry techniques, respectively.
References:
- Friedenreich CM. Physical activity and breast cancer: Review of the epidemiologic evidence and biologic mechanisms. Vol. 188, Recent Results in Cancer Research. Springer, Berlin, Heidelberg; 2011. p. 125–39.
- Harbeck N, Gnant M. Breast cancer. Vol. 389, The Lancet. 2017
Interested students are welcome to contact : [email protected]
Cell Segmentation of adipose cells tissue by advanced image-analysis techniques

Current epidemiological evidence suggests an increased relative risk for estrogen receptor-positive breast cancer. To gain important insights into the interplay between human breast epithelial cells and the surrounding stroma tissue in response to testosterone, we aim to analyze the cell distribution on the white adipose tissue after staining. In particular, significant alterations in the shape and size of the adipocytes could reveal metabolic changes that promote excessive growth of human epithelial cells.
The goal of the project is to develop an image-analysis method able to identify each adipose cell in the stroma tissue based on its distinctive visual feature—the dim membrane, which is often subtle and difficult to detect locally. Various strategies will be explored, including enhancement filters and deep learning techniques, to facilitate cell segmentation. A global analysis of the whole image will be required to make robust the cell segmentation of the matrix.
The final software package to be developed, preferably in Java / Fiji, should be versatile enough to be applicable in other contexts where images feature membranes that form a tessellation of objects.
This project is a collaboration with the EPFL Center for Imaging and the EPFL Brisken Lab.
Supervised by: [email protected], [email protected], [email protected]
Interested students are welcome to contact : [email protected]
Analysis of the Spatial Organisation of Nuclei in Human Breast Epithelium
Current epidemiological evidence suggests an increased relative risk for estrogen receptor-positive breast cancer. To gain important insights into the interplay between human breast epithelial cells and the surrounding stroma tissue in response to testosterone, we aim to analyze the cell distribution on the white adipose tissue after staining. In particular, significant alterations in the shape and size of the adipocytes could reveal metabolic changes that promote excessive growth of human epithelial cells.
The goal of the project is to develop an image-analysis method able to identify each adipose cell in the stroma tissue based on its distinctive visual feature—the dim membrane, which is often subtle and difficult to detect locally. Various strategies will be explored, including enhancement filters and deep learning techniques, to facilitate cell segmentation. A global analysis of the whole image will be required to make robust the cell segmentation of the matrix.
The final software package to be developed, preferably in Java / Fiji, should be versatile enough to be applicable in other contexts where images feature membranes that form a tessellation of objects.
References:
- Scabia V, Ayyanan A, De Martino F, Agnoletto A, Battista L, Laszlo C, et al. Estrogen receptor positive breast cancers have patient specific hormone sensitivities and rely on progesterone receptor. Nat Commun. 2022;13:3127.
- Shamseddin M, De Martino F, Constantin C, Scabia V, Lancelot A-S, Laszlo C, et al. Contraceptive progestins with androgenic properties stimulate breast epithelial cell proliferation. EMBO Mol Med. 2021;13:e14314.
This project is a collaboration with the EPFL Center for Imaging and the EPFL Brisken Lab.
Supervised by: [email protected], [email protected], [email protected]
Interested students are welcome to contact : [email protected]
