Our goal is to reveal the biophysical principles underlying the dynamic organization of cellular organelles and protein assemblies. We are fascinated by mitochondrial fission, fusion, growth, transport, and network topology, and how they contribute to energy production and mitochondrial genome maintenance.
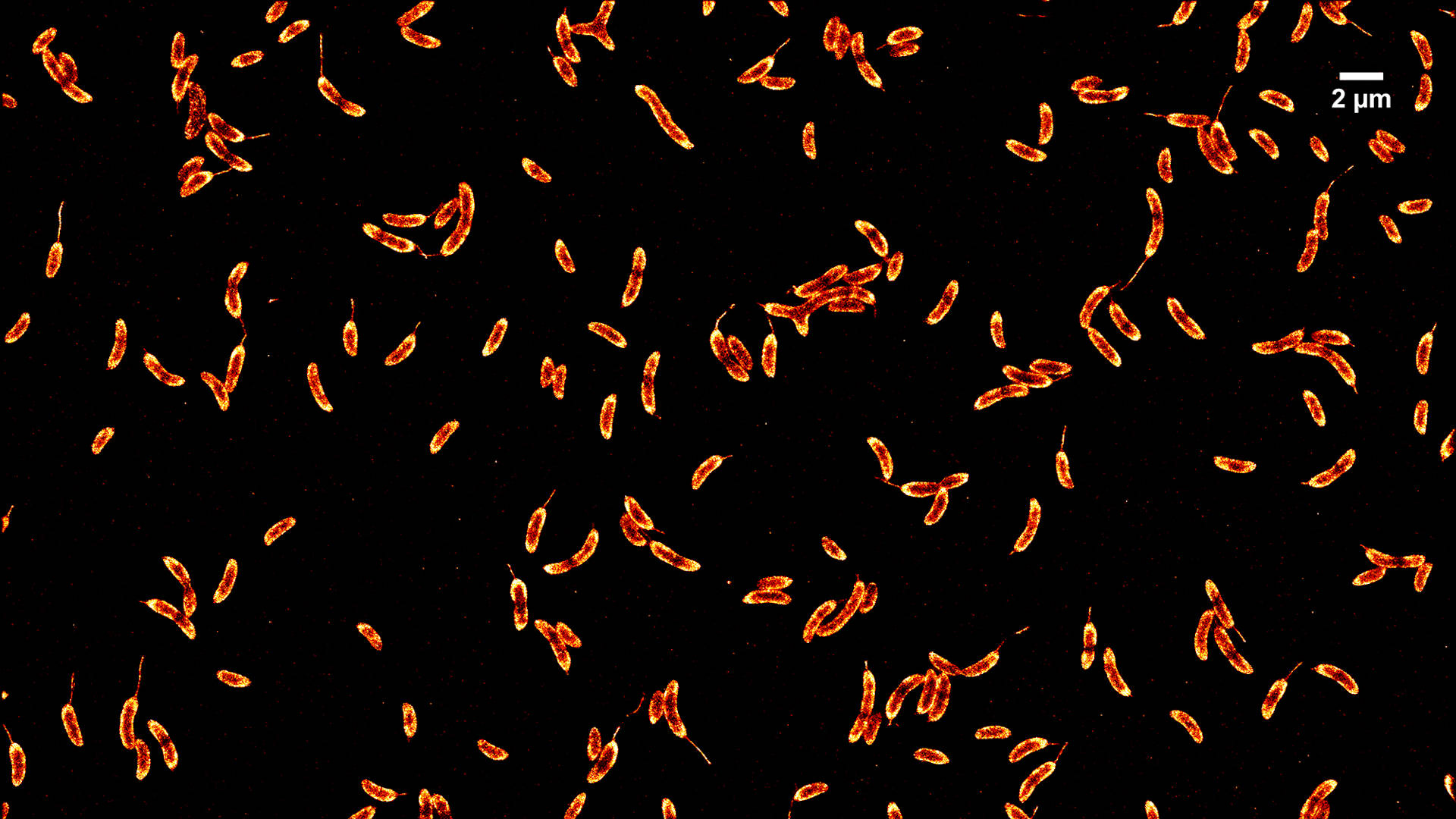
Mitochondria move along the microtubule cytoskeleton, which is organized around the centrosome: we investigate its structure and the role of post-translational modifications. Mitochondria are believed to have originated as an ancient bacterial endosymbiont: in comparison, we are also curious about how genome replication, cell growth, and division are coordinated in bacteria.
Our scientific approach is enabled by technology; we custom-build high throughput super-resolution microscopes. Enlarged fields of view, combined with automation, allow us to go beyond anecdotal images to large-scale quantification and model building. Currently, we are expanding our repertoire by integrating computer vision into our microscopes, as well as employing label-free imaging methods.
Projects
Large field-of-view super-resolution microscopy
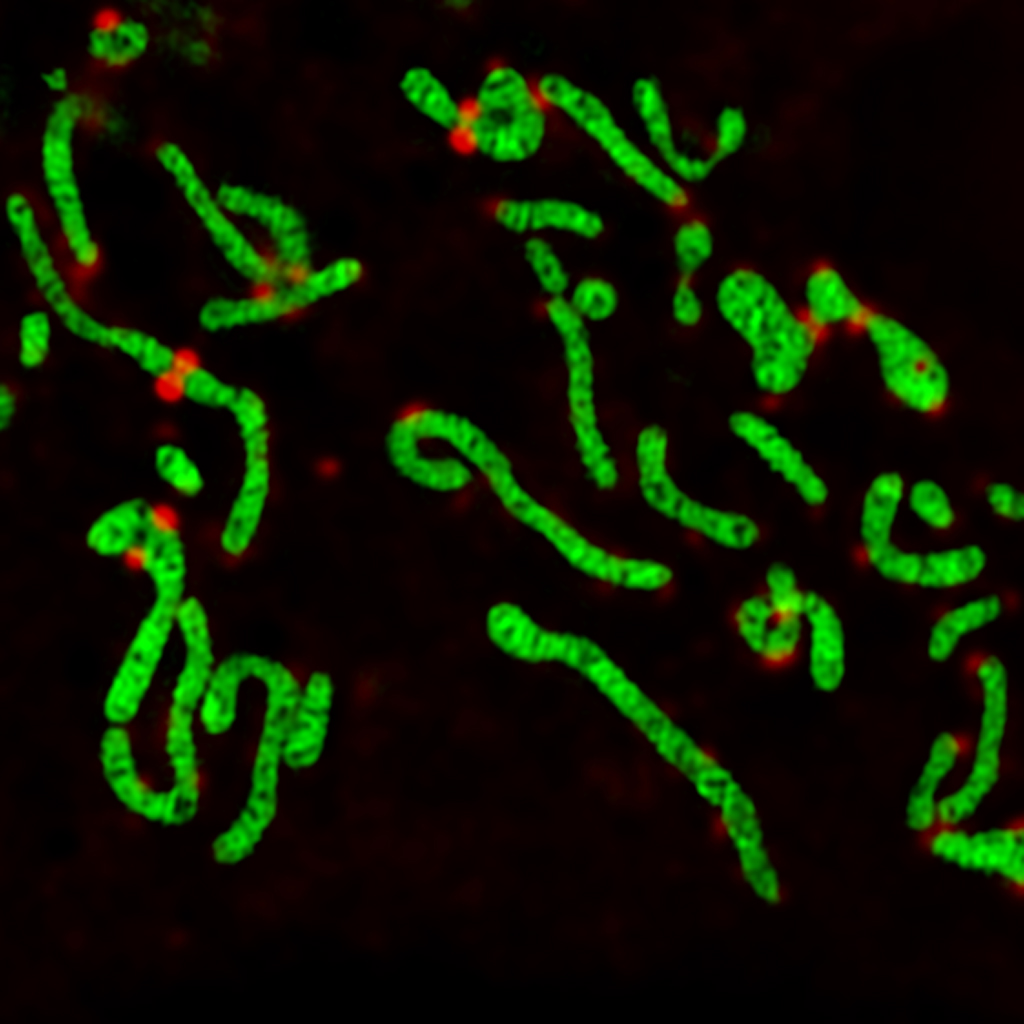
Single molecule localization microscopies (SMLM) have been established as important tools for studying cellular features with resolutions on the order of ~10 nm. We have developed imaging modalities to perform large field-of-view SMLM using microlens array (MLA)-based flat-field epi-illumination. Our system can efficiently and homogeneously illuminate a large sample area (~150×150 µm2) enabling high-throughput SMLM imaging.
Multicolor 3D single particle reconstruction
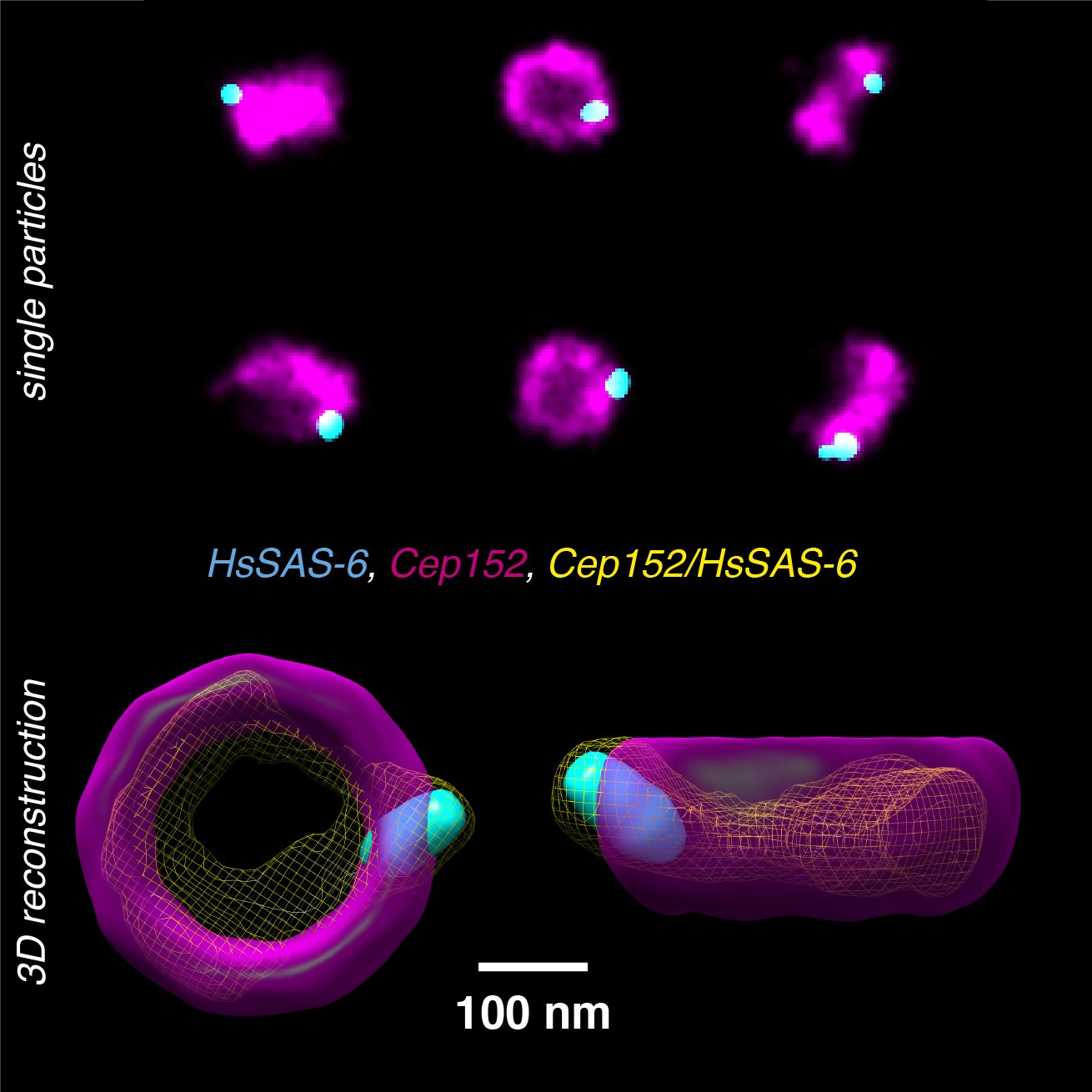
Single-particle reconstruction from electron microscopy (EM) images is today able to reach atomic resolution. However, since the image contrast in EM is dependent upon the local electron density within biological structures, the resulting image lacks direct information on protein identity. We are developing computational and analytical frameworks to reconstructs and co-align multiple proteins from multicolor 2D SMLM images.
Waveguide TIRF for high-throughput DNA-PAINT
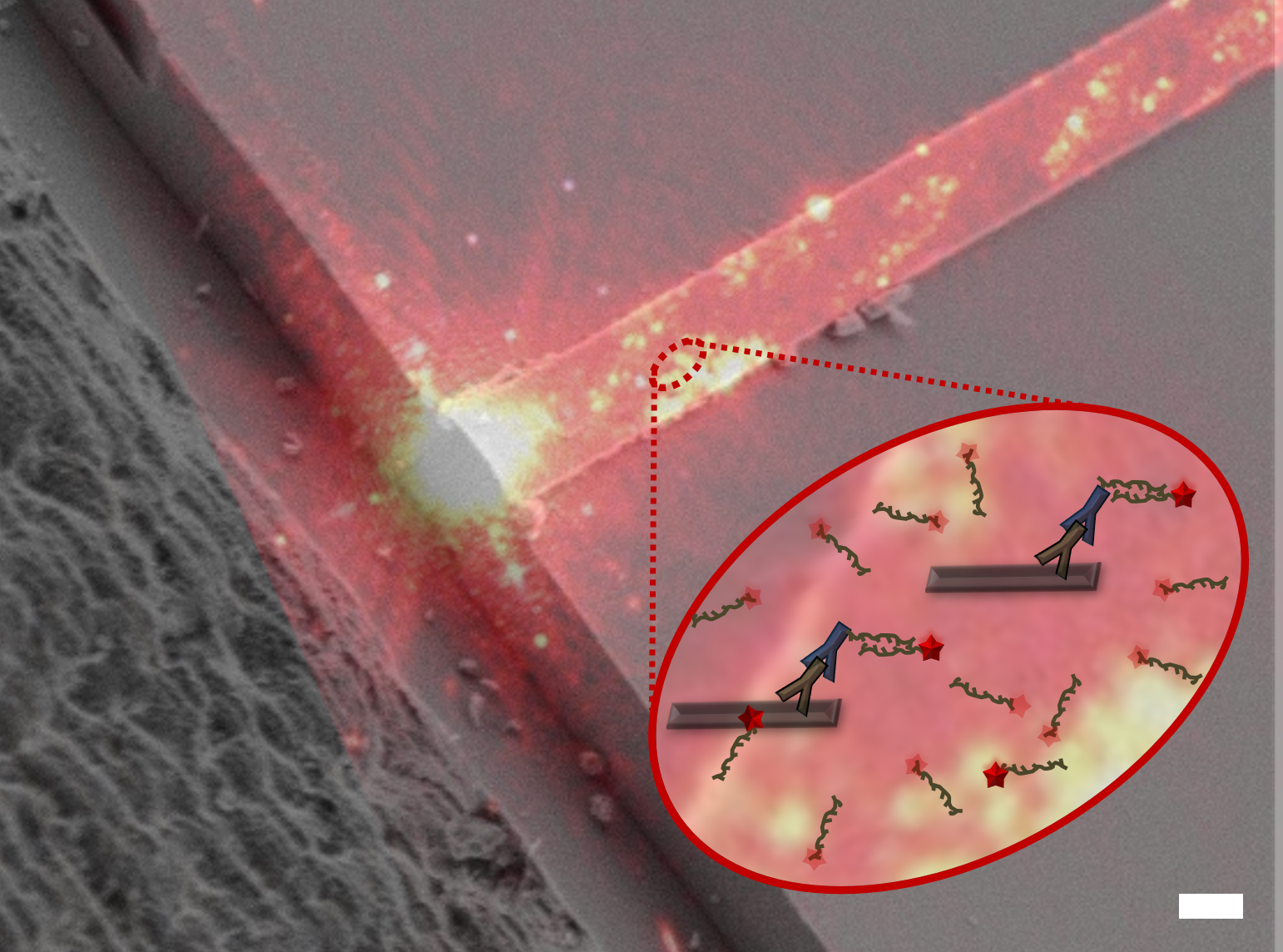
The PAINT method for localization microscopy offers higher localization precision and more continuous target sampling than standard photoswitching methods. We are using waveguides to increases throughput and data quality for PAINT. The waveguide chip is a multi-layer structure on a silicon substrate with dedicated sample wells to enable highly uniform ~100×2000 µm2 area evanescent field for TIRF illumination.
Mitochondrial dynamics
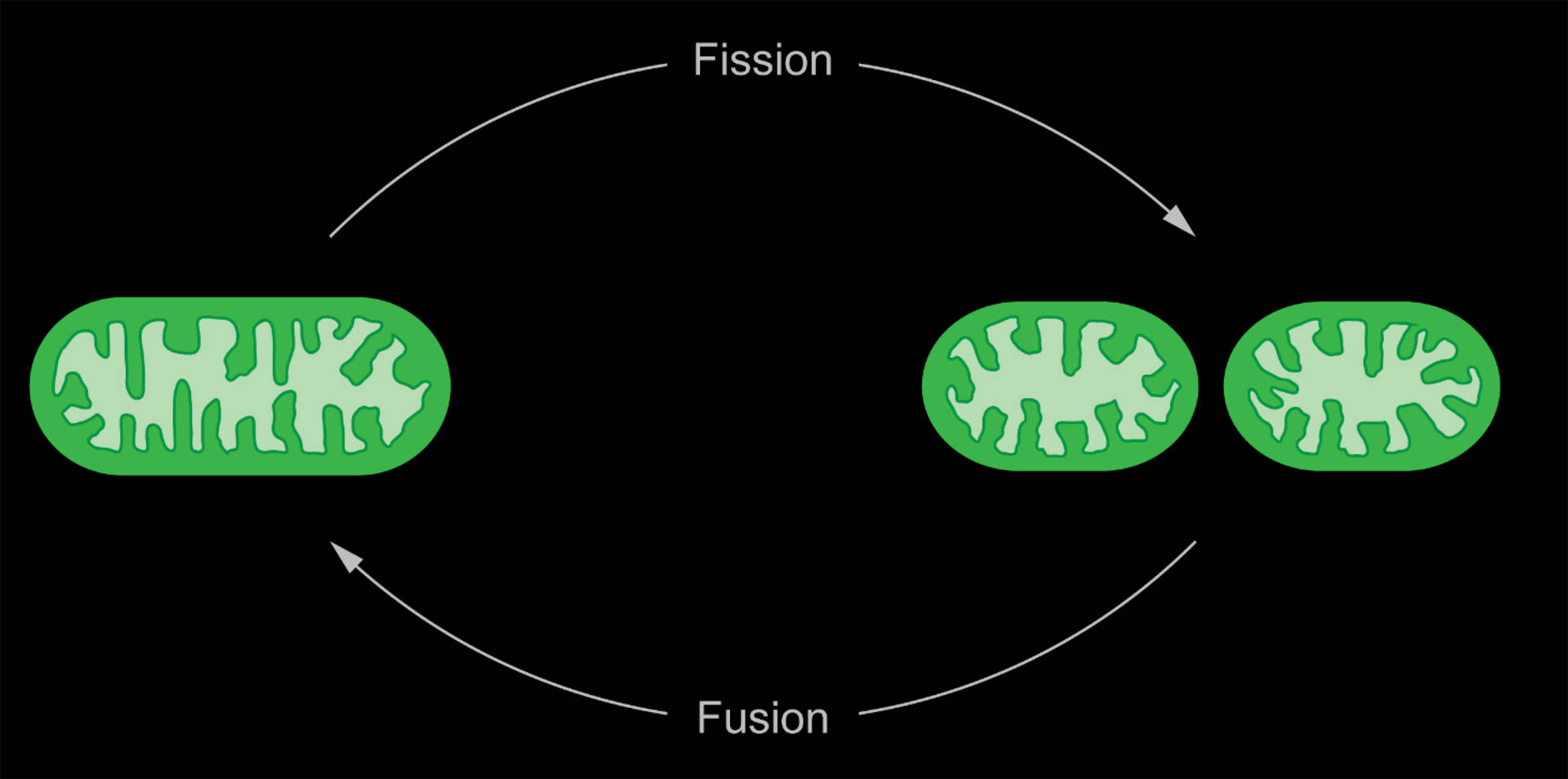
Mitochondria divide and fuse dynamically, and our aim is to unravel the physical and physiological signatures of these processes. By quantifying the dynamic geometry of mitochondrial constrictions, we extract the energies and forces required for division and link them to interactions with the division machinery. Thereby, we can develop a physical framework accounting for the energetic requirements for fission. Furthermore, we are investigating the metabolic and bioenergetic changes associated with mitochondrial division on a single-organelle level. A custom built instant-SIM enables us to perform fast, multicolor imaging on whole cells.