1. ScientificRAG: An AI Agent Framework for DW-MRI Literature Mining and Automated Computational Experimentation
 Project Context Retrieval-Augmented Generation (RAG)[4] and intelligent agents[5] offer new approaches for researchers to interact with scientific literature. These AI systems can process collections of academic papers, extract relevant information, and provide contextual answers while integrating with specialized computational tools. This technology has the potential to streamline literature review and computational analysis workflows, making them more efficient for researchers.
Project Context Retrieval-Augmented Generation (RAG)[4] and intelligent agents[5] offer new approaches for researchers to interact with scientific literature. These AI systems can process collections of academic papers, extract relevant information, and provide contextual answers while integrating with specialized computational tools. This technology has the potential to streamline literature review and computational analysis workflows, making them more efficient for researchers.
Project Overview: Develop an intelligent scientific research assistant that combines RAG capabilities with agent-based tool integration for diffusion MRI analysis. The system will ingest and index scientific papers on diffusion-weighted MRI, enabling researchers to query the literature naturally while seamlessly accessing computational tools like CACTUS[1] for substrate generation and MC-DC[2] for Monte Carlo simulations. The agent will understand research contexts, provide evidence-based answers, and automatically invoke appropriate computational tools in response to user queries.
Learning Outcomes
- RAG Systems: Master document ingestion, vector storage, and retrieval pipelines for scientific literature
- Agent Architecture: Build intelligent agents that can reason about research questions and select appropriate tools
- Scientific Computing Integration: Connect AI systems with specialized research tools (CACTUS, MC-DC) via Model Context Protocol (MCP)[3]
- DW-MRI Domain Knowledge: Develop understanding of diffusion-weighted MRI through AI-assisted literature exploration
- Production AI Systems: Create robust, scalable systems for research environments
This project consists of 50% AI/RAG coding, 40% research domain exploration, and 10% integration of computational tools, providing hands-on experience with advanced AI research tools.
Requirements
- Strong Python programming
- Interest in natural language processing and information retrieval
- Basic understanding of scientific computing workflows
- Knowledge in DW-MRI/biomedical imaging (advantageous)
- Curiosity about AI-assisted research methodologies
Technical Outcomes
- Production-ready RAG system for scientific literature analysis
- Intelligent agent capable of literature-informed computational tool selection
- Integration with CACTUS and MC-DC via MCP for seamless research workflows
- Documentation and deployment guidelines for research environments
- Demonstrated expertise in modern AI research assistance technologies
Project Milestones
- Phase 1 (Months 1-4): Core RAG system with document ingestion, vector storage, and basic query-answering
- Phase 2 (Months 4-6): Agent architecture with integration of computational tool (CACTUS, MC-DC)
Supervisors: Dr Juan Luis Villarreal ([email protected]), Dr Jonathan Rafael Patiño ([email protected]), and Prof. Jean-Philippe Thiran
[1] Villarreal-Haro et al. “CACTUS: a computational framework for generating realistic white matter microstructure substrates.” Front Neuroinform, 2023.
[2] Rafael-Patino et al. “Robust Monte-Carlo simulations in diffusion-MRI: Effect of the substrate complexity and parameter choice on the reproducibility of results.” Front Neuroinform, 2020.
[3]Singh, Aditi, et al. “A survey of the model context protocol (mcp): Standardizing context to enhance large language models (llms).” (2025).
[4]Gupta, Shailja, Rajesh Ranjan, and Surya Narayan Singh. “A comprehensive survey of retrieval-augmented generation (rag): Evolution, current landscape and future directions.” arXiv preprint arXiv:2410.12837 (2024).
[5]Sapkota, Ranjan, Konstantinos I. Roumeliotis, and Manoj Karkee. “Ai agents vs. agentic ai: A conceptual taxonomy, applications and challenges.” arXiv preprint arXiv:2505.10468 (2025).
2. Synthetic Stroke Lesion Simulation and Deep Learning Segmentation Using HCP Data
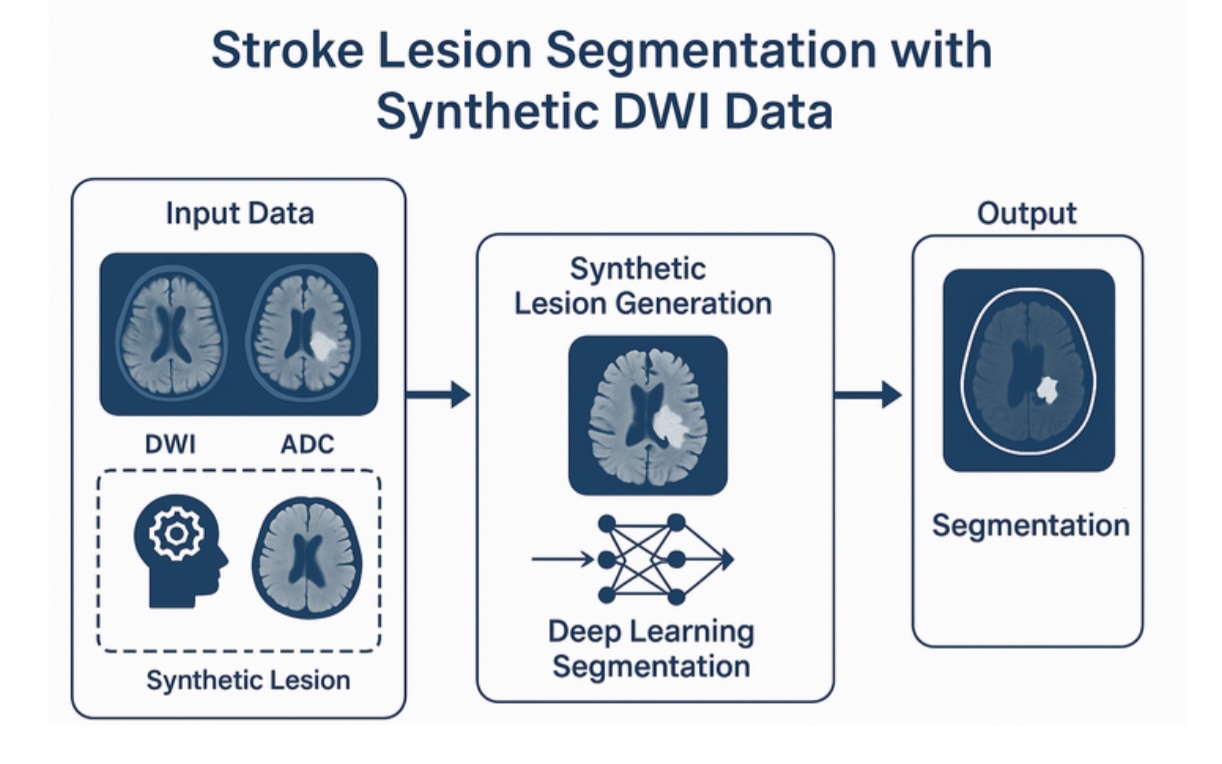
Automatic segmentation of ischemic stroke lesions in diffusion MRI is a critical task for clinical decision support. However, current supervised learning models require large and annotated datasets of stroke patients, which are difficult to obtain due to privacy concerns and annotation costs.
In this project, the student will develop a novel simulation-based framework to generate realistic synthetic acute stroke lesions using high-quality structural and diffusion MRI from the Human Connectome Project (HCP). From the high image quality of HCP subjects, synthetic lesions will be created by simulating changes in the MRI contrast inspired by known patterns of cytotoxic edema and white matter disruption. Additionally, tractography can be used to model anatomically consistent lesion propagation patterns in white matter. This dataset will be used to fully train a deep segmentation network from scratch.
The final part of the project will involve evaluating the trained model on real-world stroke imaging data from open clinical datasets, such as the ISLES challenge cohort, as well as other network prototypes developed and deployed at CHUV Lausanne University Hospital.
Requirements:
- Experience with Python and machine learning libraries (PyTorch, TensorFlow, or similar).
- Interest in diffusion MRI and image processing.
- Knowledge of neuroimaging tools (e.g., Dipy, MRtrix) is a plus.
Outcomes:
The expected outcome is a proof-of-concept segmentation pipeline trained purely on simulated data with strong performance on real-world stroke images. The student will be encouraged to co-author a scientific publication or extended abstract for submission to a machine learning or medical imaging conference (e.g., MICCAI, ISBI, MIDL).
Supervisors: Dr. Jonathan Rafael Patiño ([email protected]) and Prof. Jean-Philippe Thiran.
References:
[1] Radiopaedia. “Diffusion-weighted imaging.” Overview of DWI techniques and ADC mapping, foundational for understanding synthetic DWI/ADC in neuroimaging and lesion simulation. https://radiopaedia.org/articles/diffusion-weighted-imaging-2
[2] Sahoo P, et al. “Synthetic apparent diffusion coefficient for high b-value diffusion weighted MRI in Prostate.” This study demonstrates that ADC values for higher b-value DWI can be computed from lower b-values using a log-linear relationship, supporting the use of synthetic ADC for lesion simulation and optimized imaging contrast.
[3] “Robust Monte-Carlo Simulations in Diffusion-MRI: Effect of the Substrate Complexity and Parameter Choice on the Reproducibility of Results” (Rafael-Patino et al., 2020)
3. ProjectPyTorch Based Signal Computation Wrapper for Monte Carlo Diffusion MRI Simulation

Monte Carlo simulations are a powerful method for modeling diffusion-weighted MRI (DWI) signals based on realistic tissue microstructure. The MC/DC (Monte Carlo / Diffusion Collision) simulator is a high-performance C++/CUDA-based tool developed in-house to simulate diffusion signals from complex substrates using particle-based approaches. However, integration with modern machine learning workflows remains limited due to the lack of native bindings for optimization and neural inference.
In this project, the main task is to design and implement a PyTorch-compatible module for DWI signal computation based on stored particle trajectories generated by the MC/DC simulator. The implementation will support arbitrary MRI encoding protocols and compute synthetic diffusion signals directly from raw particle motion and collision information.
The student will develop the forward model in PyTorch, enabling direct integration into gradient-based optimization pipelines and machine learning frameworks. While the focus will be on accurate and efficient signal computation, this foundation will enable future inverse modeling and neural surrogate training. White matter microstructure substrates will be used as a testbed for the implementation.
Requirements:
- Strong experience with Python and PyTorch.
- Familiarity with C++ or CUDA is a plus but not mandatory.
- Interest in simulation-based modeling and numerical methods.
Outcome:
The final deliverable will be a validated and documented PyTorch module to be integrated into the official open-source MC/DC GitHub repository.
Supervisors: Dr. Jonathan Rafael Patiño ([email protected]) and Prof. Jean-Philippe Thiran.
References:
[1] “Robust Monte-Carlo Simulations in Diffusion-MRI: Effect of the Substrate Complexity and Parameter Choice on the Reproducibility of Results” (Rafael-Patino et al., 2020
[2] ReMiDi (Khole et al., 2025) Reconstruction of Microstructure Using a Differentiable Diffusion MRI Simulator . https://arxiv.org/abs/2502.01988
4. Exploring age-related changes in cerebral white-matter streamlines at the segment-level in youth using open-access neuroimaging data
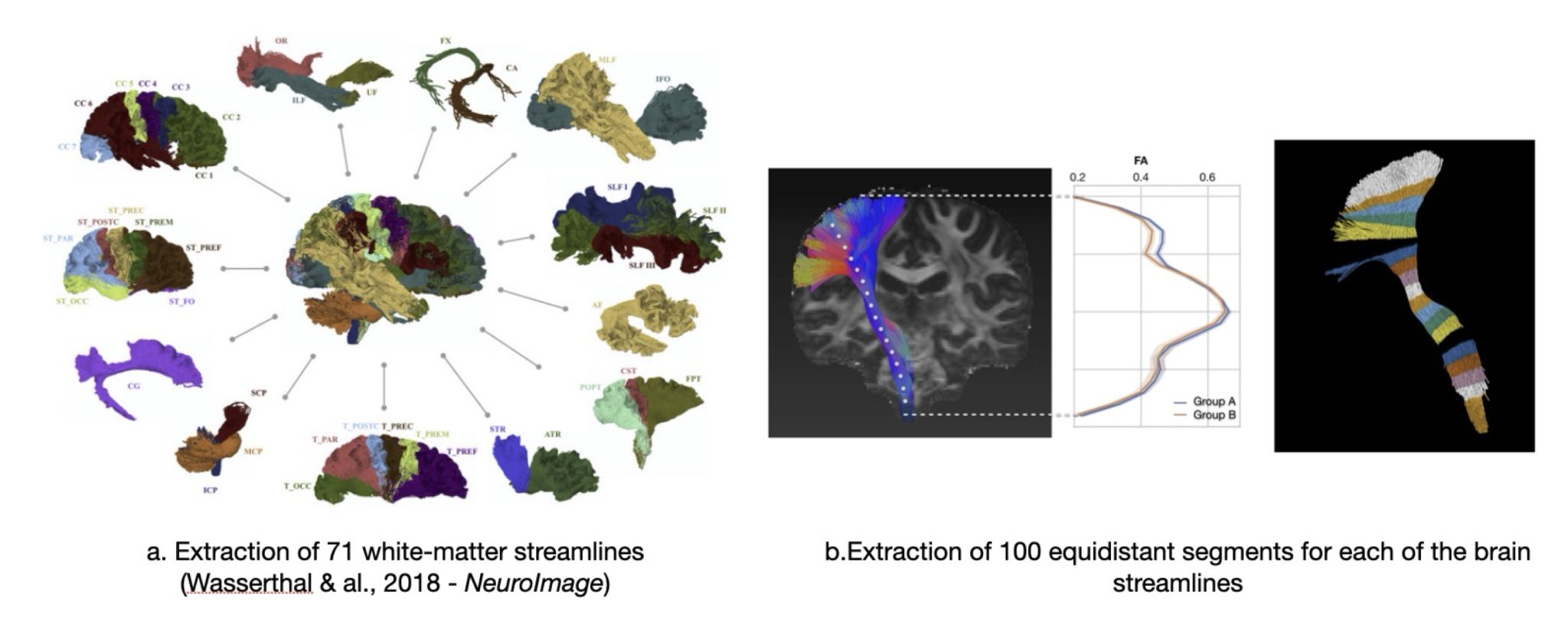
Figure. Illustration of the methods of Wasserthal et al., 2018
Background & Research Question:
Diffusion-weighted imaging enables in vivo mapping of white matter streamlines in the human brain, providing insights into how structural connectivity develops during childhood and adolescence. Previous studies (e.g., Reynolds et al., 2019, NeuroImage) have shown significant changes in white matter microstructure across this developmental period. However, these changes are typically assessed as averages across entire streamlines, which may obscure important regional variations along the streamlines.
In this project, we aim to go beyond whole-streamlines averages and investigate how white matter properties evolve along the length of individual streamlines. This fine-grained approach may reveal distinct developmental patterns occurring at different streamline locations, which could be linked to underlying neurobiological processes and cognitive outcomes.
In the context of a master project, a second stage will involve the investigation of the segment cauterization and correlation to the previously identified patterns. The student will be responsible to design and develop tailored strategies to perform tract specific segmentation of the brain streamlines.
Dataset:
We will use data from the Philadelphia Neurodevelopmental Cohort (PNC) (Satterthwaite et al., 2016), a large, publicly available dataset that includes neuroimaging and behavioral data from 700 participants with typical brain development aged 8 to 21 years.
Key features of the dataset for this project:
- Cross-sectional diffusion-weighted MRI data
- Preprocessed single-shell diffusion images
- Tractography-based segmentation using TractSeg (Wasserthal et al., 2018, NeuroImage; see Figure above) identifying 71 major white matter tracts
- Quantitative diffusion metrics—fractional anisotropy (FA) and mean diffusivity (MD)—sampled across 100 equidistant segments per tract
Objectives of the Project:
- Analyse how white matter properties (FA, MD) change with age along different segments of white matter tracts
- Identify segment-specific developmental patterns that may not be visible when averaging across whole tracts
- Apply appropriate statistical or computational approaches, such as functional data analysis or machine learning, to model these relationships
Requirement:
- Basic knowledge of statistics
- Interest in brain development and neuroimaging
- Motivation apply data analysis techniques (e.g., Python, R, or similar tools)
Supervision: Dr Jonathan Rafael Patiño ([email protected]), Vanessa Siffredi (UNIL/CHUV) and Prof. Jean-Philippe Thiran
5. Understanding brain’s White Matter organization and plasticity in children with AgCC using MRI tractography
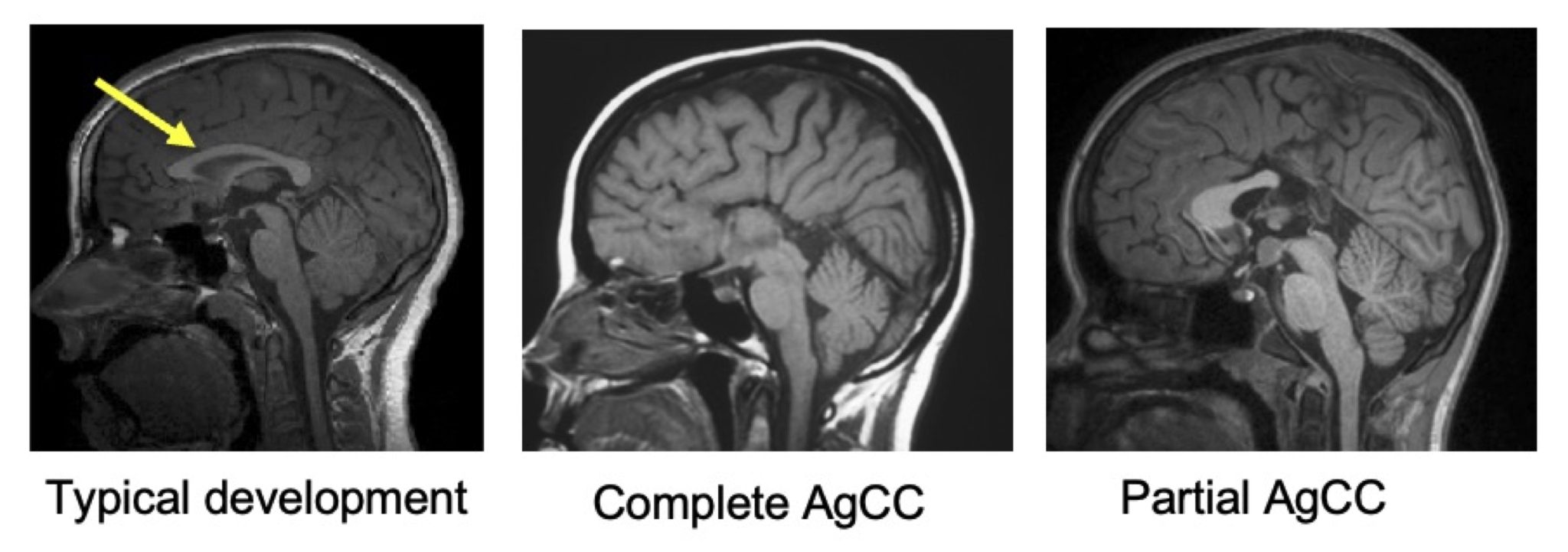
Figure. T1-weighted, sagittal slice, of typically developing children, children with complete agenesis of the corpus callosum (AgCC) and partial AgCC. The corpus callosum is indicated with a yellow arrow in the typical development case.
Background & Research Question:
The corpus callosum is the largest white matter structure in the human brain, containing over 190 million axons that connect the left and right hemispheres. A developmental absence of this structure—known as agenesis of the corpus callosum (AgCC)—is one the most common congenital brain malformation (see Figure). AgCC can result in either complete or partial absence of the corpus callosum and is associated with variable cognitive, behavioral, and neurological outcomes. Diffusion-weighted imaging enables the in vivo reconstruction of white matter pathways. This neuroimaging tool is particularly well suited for studying populations with abnormalities of white-matter development —such as AgCC—as it enables the exploration of potential structural neuroplastic responses in such atypically developing brains. Despite its clinical relevance, research in this area remains limited, largely due to the rarity of the condition. A pioneering study by Bénézit et al. (2015) explored white matter reorganization in a small sample (n = 6, https://doi.org/10.1016/j.cortex.2014.08.022).
This project aims to replicate and extend the findings of Bénézit et al. using a larger and richer dataset, with the goal of better understanding white matter organization and plasticity in children with AgCC.
Dataset:
We will use data from a unique dataset, the “Pediatric Agenesis of the Corpus Callosum Project”, collected at the Royal Children’s Hospital in Melbourne, Australia (Siffredi et al., 2021 – 10.1093/cercor/bhaa289), which contained diffusion-weighted imaging of children with AgCC (n = 20) and typically developing children (n = 29) aged 8 to 16 years.
Key features of the dataset for this project:
- Cross-sectional diffusion-weighted MRI data of a unique clinical population
- Preprocessed two-shell diffusion images
- Whole brain tractography available
- Extensive neurobehavioral and clinical assessments
Objectives of the Project:
- Replicate the key findings of Bénézit et al. (2015) which will serve as a foundational framework for the initial analysis of white-matter streamlines in this population.
- Extend the analysis, depending on student interest, in one or more of the following directions:
- Apply and compare different diffusion approaches to better characterise white-matter streamlines in such atypical brains
- Assess symmetry and asymmetry in white matter streamlines across hemispheres
- Explore atypical streamlines (e.g., sigmoid bundle)
- Explore associations with clinical outcomes
Requirement:
- Basic knowledge of image processing
- Interest in neuroimaging and clinical neuroscience
Supervision: Dr Jonathan Rafael Patiño ([email protected]), Vanessa Siffredi (UNIL/CHUV) and Prof. Jean-Philippe Thiran
6. IMU-based motion tracking and 3D modeling of the lumbosacral curvature

Problem encountered: In medical environments, infrared-based cameras are typically used for highly accurate motion tracking. These systems, however, are susceptible to occlusions, which can restrict their performance and may be unavoidable in certain use cases. A typical example is the inability to track lumbosacral curvature when a patient must lie on her back.
Project goal:Implement an alternative motion tracking technology, based on wearable inertial motion units (IMUs), for integration into a cutting-edge medical device for obstetrical care.
Project Task: Familiarize with IMUs, modelize in 3D a simplified representation of the lumbosacral curvature. Develop post-processing methods to reduce sensors-related noise and improve accuracy of the lumbosacral curvature prediction.
Requirements:
- Experience in Python / Matlab
- Experience in 3D vision and modelization
- Experience in signal and image processing
- Experience with mathematical optimization techniques
- Interest in biomotion
- Interest in wearable sensors
Outcome: A 3D reconstruction pipeline to render the sacrum position in 3D relative to other pelvic bones in real-time based on IMU sensors data and inputs from our current prototype.
Note: We are looking for a committed and dedicated student to join our journey. Integrated into a larger startup project, this PDM gives you the chance to develop impactful technology and potentially explore future career opportunities with our team.
Supervisors: Dr. Johann Hêches, Ms. Sandra Marcadent ([email protected]) and Prof. Jean-Philippe Thiran.
References:
https://www.mdpi.com/1424-8220/25/19/5963
https://openstax.org/books/anatomy-and-physiology-2e/pages/7-3-the-vertebral-column
7. Dataset Fingerprinting for Fetal Brain MRI Super-Resolution – Collaboration with CIBM SP UNIL-CHUV [Master thesis]

Fetal brain MRI is particularly vulnerable to motion, variable acquisition protocols, and heterogeneous clinical environments. These factors produce substantial differences across datasets, which can significantly impact downstream tasks such as segmentation and quality control (QC) [1]. Understanding and characterizing these distributional differences is a crucial step toward building robust and generalizable medical imaging pipelines.
This project focuses on developing dataset fingerprinting and dataset distance metrics tailored to fetal brain MRI super-resolution (SR) data. The goal is to design dataset representations—either hand-crafted embeddings derived from our existing framework FetMRQCSR [2] or learned embeddings—that capture meaningful distributional properties of a dataset. These fingerprints will then be used to compute inter-dataset distances, providing a principled way to quantify how similar or different two fetal MRI datasets are.
Such dataset distances have strong potential for data-adaptive model selection, enabling more reliable and interpretable out-of-domain generalization of machine learning models. As part of the project, the student will have the opportunity to validate how these dataset distances improve QC model selection in fetal brain MRI.
Requirements:
- Strong programming skills in Python and PyTorch
- Solid foundation in machine learning
- Interest in learning the fundamentals of fetal brain MRI and medical imaging
Outcomes:
The project aims to deliver a ready-to-use dataset fingerprinting method that will enable:
- Exploration and characterization of a large dataset of 1000+ fetal brain MRI scans,
- More efficient selection and training of generalizable ML models for fetal brain MRI quality control.
References:
[1] Zalevskyi, Vladyslav, et al. “Advances in Automated Fetal Brain MRI Segmentation and Biometry: Insights from the FeTA 2024 Challenge.” arXiv preprint arXiv:2505.02784 (2025).
[2] Sanchez, Thomas, et al. “Automatic quality control in multi-centric fetal brain MRI super-resolution reconstruction.” International Workshop on Preterm, Perinatal and Paediatric Image Analysis. Cham: Springer Nature Switzerland, 2025.
[3] Godau, Patrick, et al. “Beyond knowledge silos: Task fingerprinting for democratization of medical imaging ai.” arXiv preprint arXiv:2412.08763 (2024).
Director:
Prof. Jean-Philippe Thiran (EPFL-LTS5)
Co-supervisors:
- Dr. Thomas Sanchez CIBM SP CHUV-UNIL – Medical Image Analysis Lab ([email protected])
- Prof. Meritxell Bach CIBM SP CHUV-UNIL – Medical Image Analysis Lab ([email protected])
8. Do Foundation Models Deliver? Evaluating Data Efficiency and Generalization in Medical Imaging – Collaboration with CIBM SP UNIL-CHUV (Master Thesis)
Foundation models (FM) promise to transform medical imaging by enabling strong performance with minimal labeled data1. However, the fundamental question—whether FM fine-tuning actually outperforms training from scratch given equivalent data—remains unanswered. The first rigorous FM challenges for brain MRI (SSL3D and FOMO25, MICCAI 2025) evaluated different pre-training strategies and architectures, revealing that CNN-based approaches consistently outperformed transformers2,3. Yet these competitions benchmarked FM methods against each other, not against the from-scratch baseline. Whether pre-training provides faster convergence, better generalization, or simply comparable results to task-specific training remains an open empirical question.
This project will systematically investigate whether FM fine-tuning provides faster convergence, better generalization, or both, compared to training equivalent architectures from scratch. Using our challenge-winning CNN-based foundation models2, the student will design controlled experiments varying: (1) training set size (e.g., 10, 50, 100, 200 samples), (2) task type (segmentation vs. classification), and (3) clinical domain (Multiple Sclerosis lesion segmentation across different MRI sequences and resolutions, and fetal brain tissue segmentation at varying gestational ages). The goal is to characterize under what data regimes FM advantages emerge, persist, or disappear.

Multiple Sclerosis: MRI with automated lesion prediction overlay in yellow. Fetal MRI and brain tissue segmentation.
Requirements:
- Strong programming skills in Python and PyTorch
- Solid foundation in machine learning and deep learning
- Interest in experimental design and medical imaging
Outcomes:
- Empirical characterization of FM benefits across data regimes, tasks, and imaging domains
- Practical guidelines for when FM fine-tuning outperforms training from scratch
- Reproducible benchmarking framework applicable to future FM evaluation
References:
[1] Moor, M. et al. Foundation models for generalist medical artificial intelligence. Nature 616, 259–265 (2023).
[2] Gordaliza, P.M. et al. From 100,000+ images to winning the first brain MRI foundation model challenges. Under review, Nature Machine Intelligence (2025).
[3] Wald, T. et al. An OpenMind for 3D medical vision self-supervised learning. arXiv:2412.17041 (2025).
Director: Prof. Jean-Philippe Thiran (EPFL-LTS5)
Co-supervisors:
- Pedro M. Gordaliza – CIBM SP CHUV-UNIL – Medical Image Analysis Lab ([email protected])
- Meritxell Bach Cuadra – CIBM SP CHUV-UNIL – Medical Image Analysis Lab ([email protected])
9. Domain randomized pathology simulation for robust fetal brain MRI segmentation – Collaboration with CIBM SP UNIL-CHUV

Automated methods for medical image segmentations are often developed on datasets featuring mostly healthy subjects. These dataset are typically small and heterogeneous, which poses a challenge for generalization of learning-based methods. This problem is even stronger in fetal brain MRI, where the brain anatomy undergoes rapid and large changes. Recent works, based on synthetic data and domain randomization [1,2] are promising avenues for building robust models to tackle this challenge. However, they fail to generalize to pathological subjects.
In this project, we will aim at expanding our synthetic data generator to simulate pathology-like alterations. CINeMA [3] is a promising approach based on implicit neural representations that can simulate various conditions in an anatomically realistic way by interpolating them. Based on this generator, we will train a robust segmentation model that will then be tested on various pathological datasets. If time allows, we will also explore how these models could be fine-tuned on different tasks to maximize their performance and re-usability [4]. This project will provide valuable input to an ongoing research effort to characterize fetal neurodevelopment using low-field 0.55T MRI scanners, which are expected to be much more accessible to low-income countries than conventional MRI scanners.
The student will learn to : 1) handle and process 3D clinical fetal MR images, 2) learn to use state-of-the-art domain randomization techniques, 3) become familiar with state-of-the-art implicit neural representation models, 4) explore how these models could be fine-tuned to maximize their performance in related tasks. The ideal candidate for this project should have solid programming skills with proficiency in PyTorch and a strong foundation in image processing and deep learning.
References:
[1] Billot, Benjamin, et al. “SynthSeg: Segmentation of brain MRI scans of any contrast and resolution without retraining.” Medical image analysis 86 (2023): 102789.
[2] Zalevskyi, Vladyslav, et al. “DRIFTS: Optimizing Domain Randomization with Synthetic Data and Weight Interpolation for Fetal Brain Tissue Segmentation.” arXiv preprint arXiv:2411.06842 (2024).
[3] Dannecker, Maik, et al. “CINeMA: Conditional Implicit Neural Multi-Modal Atlas for a Spatio-Temporal Representation of the Perinatal Brain.” arXiv preprint arXiv:2506.09668 (2025).
[4] Wortsman, Mitchell, et al. “Model soups: averaging weights of multiple fine-tuned models improves accuracy without increasing inference time.” International conference on machine learning. PMLR, 2022.
Director:
Prof. Jean-Philippe Thiran (EPFL-LTS5)
Co-supervisors:
- Vladyslav Zalevskyi CHUV-UNIL – Medical Image Analysis Lab ([email protected])
- Thomas Sanchez CIBM SP CHUV-UNIL – Medical Image Analysis Lab ([email protected])
- Dr Meritxell Bach CIBM SP CHUV-UNIL – Medical Image Analysis Lab ([email protected])
10. Biophysical Modelling of Microstructural Diffusion MRI in the Rodent Brain – collaboration with CHUV
Project description: Diffusion MRI is a powerful tool to quantify biological tissue microstructure in vivo and non-invasively. The goal of this project is to use advanced biophysical models of diffusion MRI to extract quantitative maps of brainmicrostructure in animal models of disease vs healthy controls. Possible applications include Parkinson’s disease, creatinedeficiency or stroke. Because the estimation of these model parameters can be challenging (degeneracy or high uncertainty), the optimization of model fitting (e.g. via soft constraints or Bayesian uncertainty estimation) to improvebiological interpretability is also a possible project lead.

This project will be carried out within the Radiology Research Unit of the CHUV at Biopôle, under the supervision of Dr. RitaOliveira and Prof. Ileana Jelescu.
Start date: from January 2026 onward
Profile: We are seeking a highly motivated master’s student with a background in Electrical/Micro/Life ScienceEngineering or Physics. The project is flexible and can be adapted based on the student’s background and on the Universityprogramme’s requirements. By the end, you will be able to confidently process neuroimaging data and work withbiophysical models of brain microstructure. Very good coding skills are essential.
Interested? Contact [email protected] or [email protected]
11. Unraveling the contrast mechanism of neuromelanin-sensitive MRI – collaboration with CHUV
Project description: Neuromelanin-sensitive MRI is a novel technique that holds high clinical potential, as it detects pathological alterations in a wide range of diseases, among them Parkinson’s disease and schizophrenia. While its namesuggests that it measures the pigment neuromelanin, a surrogate of catecholaminergic function, the physical mechanismsunderlying this MRI contrast are debated in the literature. Some researchers even doubt the sequence’s sensitivity to neuromelanin at all. Understanding the underlying physics is essential for quantitative neuromelanin imaging, which could improve early diagnosis and monitoring of neurodegenerative disorders. We aim to explain
the neuromelanin-sensitive MRI contrast using computer simulations of the MRI signal. In our lab, we generate synthetic tissue using computeralgorithms to study the signal in diffusion MRI. The goal of this master’sproject is to extend our computer simulations to include two effects pivotal for the neuromelanin-sensitive MRI contrast: longitudinal relaxation and magnetization transfer.
This project will be carried out within the Radiology Research Unit of theCHUV at Biopôle, under the supervision of Dr. Malte Brammerloh and Prof. Ileana Jelescu.

What the student will deliver:
- Implement longitudinal-relaxation and magnetization-transfer modules in the existing simulation
- Validate the extended model against published
Who we are looking for:
- Strong programming skills (C++)
- Experience with numerical solvers or Monte-Carlo
- Basic knowledge of MRI physics (helpful but not mandatory).
- High motivation to model physical processes and interest in neuro-imaging.
Start date: from January 2026 onward
Why join us: The project bridges physics, computer science, and neuroscience, offering an interdisciplinary environment for students eager to contribute to cutting-edge MRI research. If you are ready to advance neuromelaninimaging and gain hands-on experience with state-of-the-art simulation tools, we invite you to apply.
Interested? Contact [email protected] or [email protected]