Reconstruction and Analysis of Cellular Networks
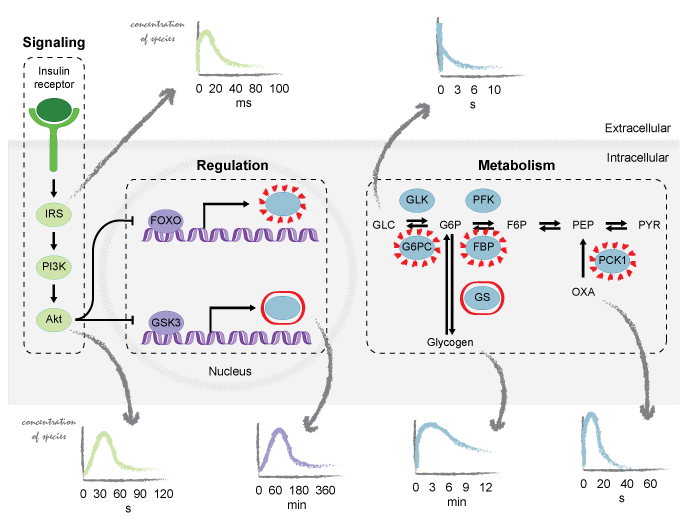 Understanding the cellular processes is crucial for making progress in medicine, biology, and biotechnology. These fields aim to characterize the behavior of the cell under different conditions to provide tools for personalized and precision medicine, green energy or efficient chemical production. Experimental approaches are currently generating an abundant amount of biological data and require computational methods to perform integrative analysis of the cellular processes.
Understanding the cellular processes is crucial for making progress in medicine, biology, and biotechnology. These fields aim to characterize the behavior of the cell under different conditions to provide tools for personalized and precision medicine, green energy or efficient chemical production. Experimental approaches are currently generating an abundant amount of biological data and require computational methods to perform integrative analysis of the cellular processes.
In our group, we focus on the reconstruction, analysis, and design of biological networks. We have developed models of metabolism, signaling, and translation. These models serve to integrate and interpret qualitative and quantitative information from cellular systems. We further tailor mathematical frameworks to identify the key elements that govern the cellular processes and to decipher their design principles. The effect of bioenergetics on the cell’s behavior stays among our primary interests.
Kinetic Modelling
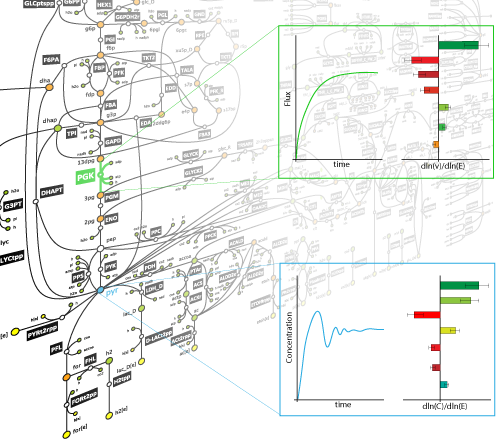 The majority of studies of metabolic processes are mainly focused on the assumption that the system operates within the time window where all system components are at steady-state. Thermodynamics-based flux balance analysis can capture the behavior of the system in these cases. However, when we seek to investigate the effect of a perturbation to the system, this assumption does not hold anymore, and the behavior of the system cannot be fully characterized only by the stoichiometric and thermodynamic properties. Such questions are mainly studied in the field of metabolic control analysis (MCA). This analysis requires that we have explicit formulations of the kinetic expressions that govern each one of the metabolic reactions. Nevertheless, there exists uncertainty in several layers, such as: network topology, formulation of kinetic rules, and parameter values. In our research we seek to build methodologies that allow us to assess the uncertainty at every layer in order to generate kinetic models that adequately describe the system and can be used to test hypotheses formulated by experiments, as well as to allow for predictions that can be further tested in experiments.
The majority of studies of metabolic processes are mainly focused on the assumption that the system operates within the time window where all system components are at steady-state. Thermodynamics-based flux balance analysis can capture the behavior of the system in these cases. However, when we seek to investigate the effect of a perturbation to the system, this assumption does not hold anymore, and the behavior of the system cannot be fully characterized only by the stoichiometric and thermodynamic properties. Such questions are mainly studied in the field of metabolic control analysis (MCA). This analysis requires that we have explicit formulations of the kinetic expressions that govern each one of the metabolic reactions. Nevertheless, there exists uncertainty in several layers, such as: network topology, formulation of kinetic rules, and parameter values. In our research we seek to build methodologies that allow us to assess the uncertainty at every layer in order to generate kinetic models that adequately describe the system and can be used to test hypotheses formulated by experiments, as well as to allow for predictions that can be further tested in experiments.
Discovery of Novel Biotransformations
 Living organisms utilize enzyme-catalyzed reactions to synthesize a large array of complex molecules. Enzyme catalyzed processes are characterized by mild conditions, fast reaction rates, highly stereospecific interactions, and minimal toxic byproduct formation. However, living organisms often consist of thousands of metabolites undergoing thousands of reactions. These reactions are carefully regulated through mechanisms developed over millions of years of evolution. An understanding of this complex system and regulation will enable the engineering of enzymes and pathways for the biosynthesis of industrial chemicals or novel pharmaceuticals. The objective of this project is the development of a computational framework for the discovery and the rational design of novel biosynthetic pathways for the production of useful or novel chemicals.
Living organisms utilize enzyme-catalyzed reactions to synthesize a large array of complex molecules. Enzyme catalyzed processes are characterized by mild conditions, fast reaction rates, highly stereospecific interactions, and minimal toxic byproduct formation. However, living organisms often consist of thousands of metabolites undergoing thousands of reactions. These reactions are carefully regulated through mechanisms developed over millions of years of evolution. An understanding of this complex system and regulation will enable the engineering of enzymes and pathways for the biosynthesis of industrial chemicals or novel pharmaceuticals. The objective of this project is the development of a computational framework for the discovery and the rational design of novel biosynthetic pathways for the production of useful or novel chemicals.