Student projects
We are constantly looking for motivated Bachelors and Masters students from any scientific or engineering background to experience interdisciplinary research with us. Here are some examples of previous and current projects, to give you an idea of the kinds of projects students do in our group. To request a future project, please go to our Members page and contact one of us directly based on your interests.
Completed projects
1) Molecular turnover in biological condensates (TP4 project, experimental biophysics and data analysis, Emilie Cuillery)
Motivation: Mitochondria are organelles which produce ATP, the energy currency of the cell, and maintain a small genome that encodes proteins required for that process. We want to investigate how mitochondrial RNA transcripts are organized.
Background: Mitochondria contain the genetic information and expression machinery to produce essential respiratory chain proteins. Within the mitochondrial matrix, newly synthesized RNA, RNA processing proteins and mitoribosome assembly factors form punctate sub-compartments referred to as mitochondrial RNA granules (MRGs). Despite their proposed importance in regulating gene expression, the structural and dynamic properties of MRGs remain largely unknown.
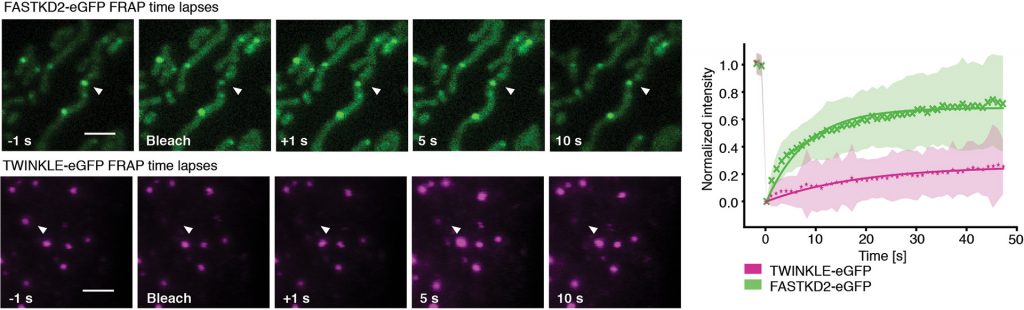
Details: We performed fluorescence recovery after photobleaching (FRAP) measurements on MRGs and mitochondrial nucleoids. A challenging part was that the mitochondria are dynamic, so they move between images. Thus, we needed to track the photobleached objects and quantify their fluorescence in each image. We developed an analysis pipeline to take this into account.
Publication: T Rey+, S Zaganelli, E Cuillery, E Vartholomaiou, M Croisier, JC Martinou and S Manley+, “Mitochondrial RNA granules are fluid condensates positioned by membrane dynamics,” Nature Cell Biology doi:10.1038/s41556-020-00584-8 (2020)
2) Automating super-resolution localization microscopy (MA project, computational microscopy, Baptiste Ottino)
Motivation: Automation allows higher quality data, and higher throughput. This is essential for quantitative microscopy, and has particularly interesting challenges in the case of super-resolution.
Background: The activation laser power in a single-molecule localization microscopy (SMLM) experiment controls the density of fluorescent molecules emitting at any given time. Precise control of the laser power is paramount; a low emitter density increases the measurement time and reduces the throughput, while an excessively high density causes the images of molecules to overlap and introduces artifacts in the recontructed image. Controlling the emitter density threfore requires expertise, as well as constant attention from the operator throughout the measurement to compensate for photobleaching.
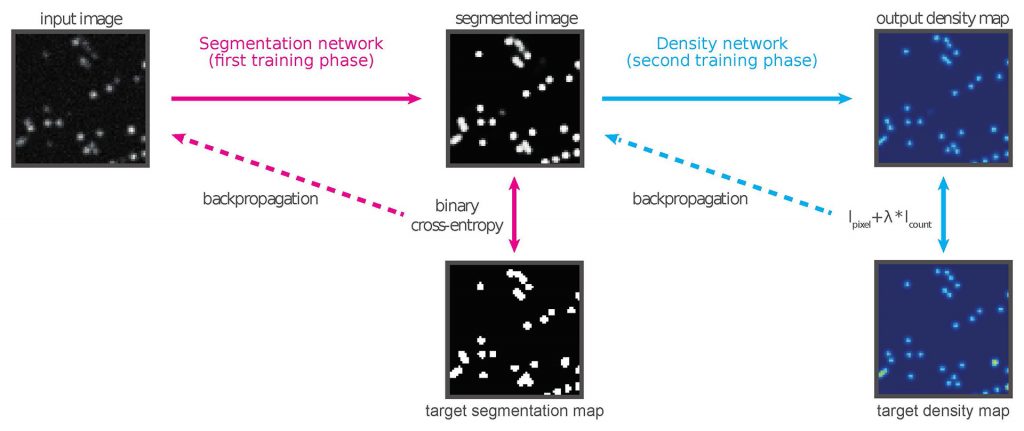
Details: We developed DEFCoN, a fully convolutional neural network that calculates the number of emitters in each frame in real time by mapping the microscopy image to a density map. This method circumvents the di cult problem of detecting individual molecules when they overlap. We demonstrate the effectiveness of this approach by comparing the counting performance of DEFCoN to the state-of-the-art detection methods for SMLM, on both simulated and real data. Taking advantage of a parallelized GPU implementation, we also demonstrate the suitability of DEFCoN for online analysis by measuring the time needed to process frames of different sizes. Finally, we demonstrate that DEFCoN can be used to give a quantitative expression of the amount of overlap in the image at any given time. We show that exploiting this property in a feedback control system is a significant step towards complete automation of the SMLM experimental process.
Publication: M Stefko, B Ottino, KM Douglass and S Manley+, “Autonomous illumination control for localization microscopy,” Opt Express doi:10.1364/OE.26.030882 (2018)
Courses
Biophysics: Physics of the Cell (PHYS-301)
To examine cellular systems through physical models and quantitative approaches to analysis.
- MINEUR, 2015, Spring semester, language : en
General Physics I
Physique générale II: Thermodynamique