Developing processes and proof of concept of the CryoNanoSIMS
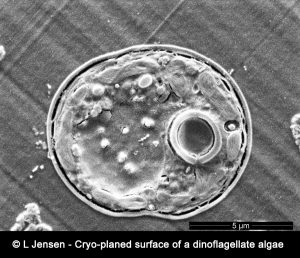
The conventional NanoSIMS instrument operates at room temperature and ultra-high vacuum (10-9-10-10 Torr). In order to preserve this vacuum, biological samples must be prepared, typically using classical sample preparation procedures developed for electron-beam imaging techniques e.g. TEM and SEM, to minimize volatilization or degassing. However, a multitude of fundamental biological processes involve the action of soluble compounds (ions, metabolites, drugs, etc.) that cannot be imaged with the conventional NanoSIMS instrument because they are lost or significantly displaced during the classical sample preparation. The only certain way to preserve and observe soluble molecular compounds and ions unperturbed in situ in a biological tissue is to create and maintain highly controlled cryo-conditions throughout the chain of preparation and analysis. Our method is based on state-of-the-art cryo-methods for sample preparation i.e. high-pressure freezing, cryo-planing, subsequent ultra-structural observations with cryo-SEM and finally NanoSIMS analysis under cryogenic conditions: CryoNanoSIMS.
Tracing and analyzing micro and nanoparticles in large tissue matrices
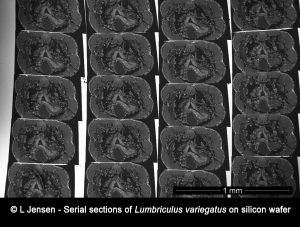
The rise in the use of engineered metal nanoparticles as well as the growing concern of microplastic pollution, has led to increased interest in tracing and analyzing such particles inside model organisms. However, such organisms pose a very large matrix compared to the minute size of the particles and localizing the particles prior to potential analysis using the NanoSIMS poses a great challenge.
In this project we use different techniques such as array tomography, reflection microscopy, scanning electron microscopy, STEM EDX and NanoSIMS to facilitate localization and analysis of particles taken up by, and distributed in whole model organisms.